You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002449_03685
You are here: Home > Sequence: MGYG000002449_03685
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides acidifaciens | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides acidifaciens | |||||||||||
| CAZyme ID | MGYG000002449_03685 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4880; End: 7963 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 30 | 720 | 5.8e-80 | 0.8058252427184466 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 1.46e-08 | 202 | 575 | 401 | 761 | alpha-L-rhamnosidase. |
| cd10277 | PQQ_ADH_I | 0.003 | 762 | 840 | 229 | 313 | Ethanol dehydrogenase, a bacterial quinoprotein (PQQ-dependent type I alcohol dehydrogenase). This bacterial family of homodimeric ethanol dehydrogenases utilize pyrroloquinoline quinone (PQQ) as a cofactor. It represents proteins whose expression may be induced by ethanol, and which are similar to quinoprotein methanol dehydrogenases, but have higher specificities for ethanol and other primary and secondary alcohols. Dehydrogenases with PQQ cofactors, such as ethanol, methanol, and membrane-bound glucose dehydrogenases, form an 8-bladed beta-propeller. |
| pfam17132 | Glyco_hydro_106 | 0.003 | 36 | 110 | 2 | 84 | alpha-L-rhamnosidase. |
| pfam08531 | Bac_rhamnosid_N | 0.004 | 940 | 975 | 17 | 63 | Alpha-L-rhamnosidase N-terminal domain. This family consists of bacterial rhamnosidase A and B enzymes. This domain is probably involved in substrate recognition. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT50138.1 | 0.0 | 1 | 1027 | 11 | 1037 |
| QKH96611.1 | 0.0 | 2 | 1027 | 12 | 1037 |
| SCM58509.1 | 0.0 | 14 | 1026 | 23 | 1029 |
| BBD46126.1 | 0.0 | 27 | 1027 | 32 | 1035 |
| QGT72310.1 | 0.0 | 17 | 1026 | 28 | 1034 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNA8 | 5.20e-11 | 30 | 1022 | 30 | 1142 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
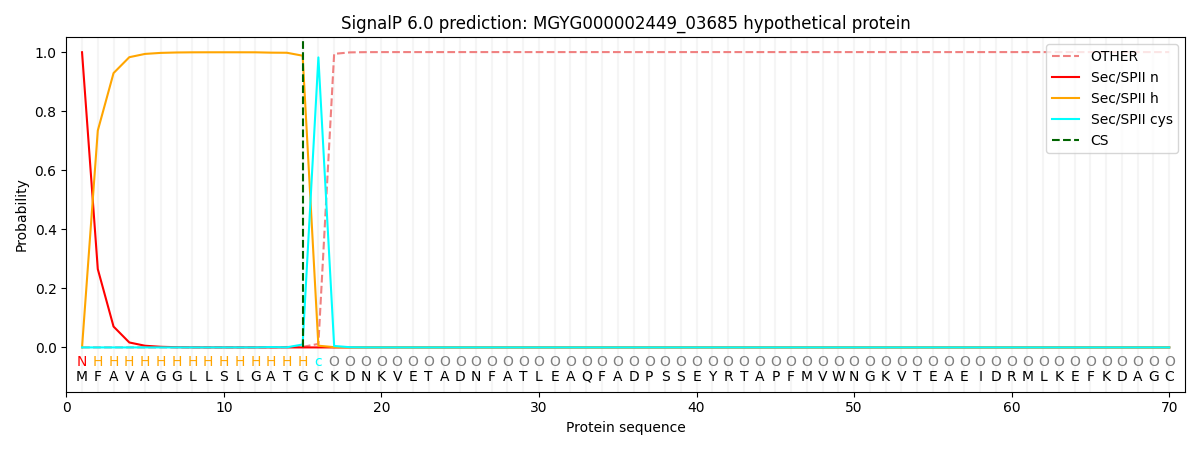
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000098 | 0.000394 | 0.999548 | 0.000000 | 0.000000 | 0.000000 |
