You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002450_01127
You are here: Home > Sequence: MGYG000002450_01127
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
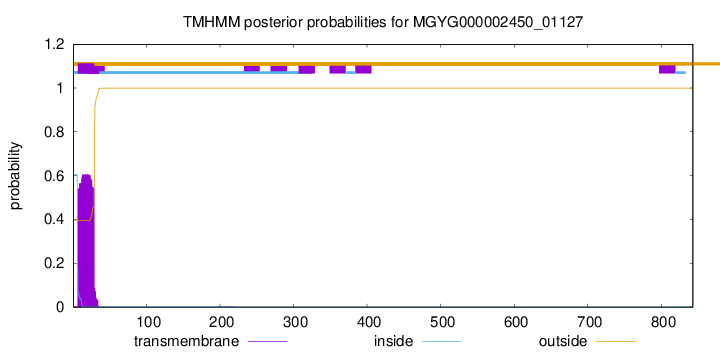
TMHMM annotations
Basic Information help
| Species | Olsenella_E sp003609875 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Coriobacteriia; Coriobacteriales; Atopobiaceae; Olsenella_E; Olsenella_E sp003609875 | |||||||||||
| CAZyme ID | MGYG000002450_01127 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1259895; End: 1262426 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 208 | 410 | 8e-26 | 0.7870370370370371 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 5.23e-14 | 191 | 522 | 54 | 358 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 4.87e-08 | 156 | 431 | 25 | 280 | Glycosyl hydrolase family 3 N terminal domain. |
| PRK15098 | PRK15098 | 2.65e-06 | 212 | 348 | 121 | 247 | beta-glucosidase BglX. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOY60939.1 | 4.80e-286 | 2 | 842 | 5 | 814 |
| BCJ97597.1 | 4.37e-178 | 46 | 836 | 70 | 843 |
| ADK68813.1 | 6.80e-168 | 2 | 837 | 4 | 820 |
| QGY45729.1 | 6.14e-149 | 43 | 832 | 34 | 832 |
| QIA06569.1 | 1.45e-148 | 43 | 832 | 34 | 827 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q46684 | 4.97e-42 | 85 | 516 | 36 | 486 | Periplasmic beta-glucosidase/beta-xylosidase OS=Dickeya chrysanthemi OX=556 GN=bgxA PE=3 SV=1 |
| Q2UFP8 | 4.01e-37 | 106 | 534 | 45 | 476 | Probable beta-glucosidase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=bglC PE=3 SV=2 |
| B8NGU6 | 6.92e-37 | 101 | 534 | 36 | 472 | Probable beta-glucosidase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bglC PE=3 SV=1 |
| Q5BCC6 | 1.53e-33 | 104 | 795 | 35 | 601 | Beta-glucosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TATLIPO

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 0.000068 | 0.000063 | 0.999851 | 0.000000 |