You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002453_01414
You are here: Home > Sequence: MGYG000002453_01414
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Akkermansia muciniphila_B | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Verrucomicrobiales; Akkermansiaceae; Akkermansia; Akkermansia muciniphila_B | |||||||||||
| CAZyme ID | MGYG000002453_01414 | |||||||||||
| CAZy Family | GH110 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 42671; End: 44461 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH110 | 94 | 588 | 2.8e-167 | 0.9087591240875912 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWP25938.1 | 0.0 | 1 | 596 | 1 | 596 |
| QWP02072.1 | 0.0 | 1 | 596 | 1 | 596 |
| QWP20708.1 | 0.0 | 1 | 596 | 1 | 596 |
| QWP52727.1 | 0.0 | 1 | 596 | 1 | 596 |
| QWP62463.1 | 0.0 | 1 | 596 | 1 | 596 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7V6M_A | 4.98e-06 | 38 | 212 | 10 | 199 | ChainA, Fibronectin type III domain-containing protein [Tyzzerella nexilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B2UL12 | 0.0 | 1 | 596 | 1 | 596 | Alpha-1,3-galactosidase A OS=Akkermansia muciniphila (strain ATCC BAA-835 / DSM 22959 / JCM 33894 / BCRC 81048 / CCUG 64013 / CIP 107961 / Muc) OX=349741 GN=glaA PE=3 SV=1 |
| Q8A2Z5 | 2.06e-92 | 98 | 568 | 45 | 518 | Alpha-1,3-galactosidase A OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=glaA PE=3 SV=1 |
| Q64MU6 | 1.55e-85 | 88 | 582 | 47 | 552 | Alpha-1,3-galactosidase A OS=Bacteroides fragilis (strain YCH46) OX=295405 GN=glaA PE=3 SV=1 |
| Q5L7M8 | 5.96e-85 | 88 | 582 | 47 | 552 | Alpha-1,3-galactosidase A OS=Bacteroides fragilis (strain ATCC 25285 / DSM 2151 / CCUG 4856 / JCM 11019 / NCTC 9343 / Onslow) OX=272559 GN=glaA PE=3 SV=1 |
| Q826C5 | 1.21e-66 | 84 | 588 | 59 | 592 | Alpha-1,3-galactosidase A OS=Streptomyces avermitilis (strain ATCC 31267 / DSM 46492 / JCM 5070 / NBRC 14893 / NCIMB 12804 / NRRL 8165 / MA-4680) OX=227882 GN=glaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
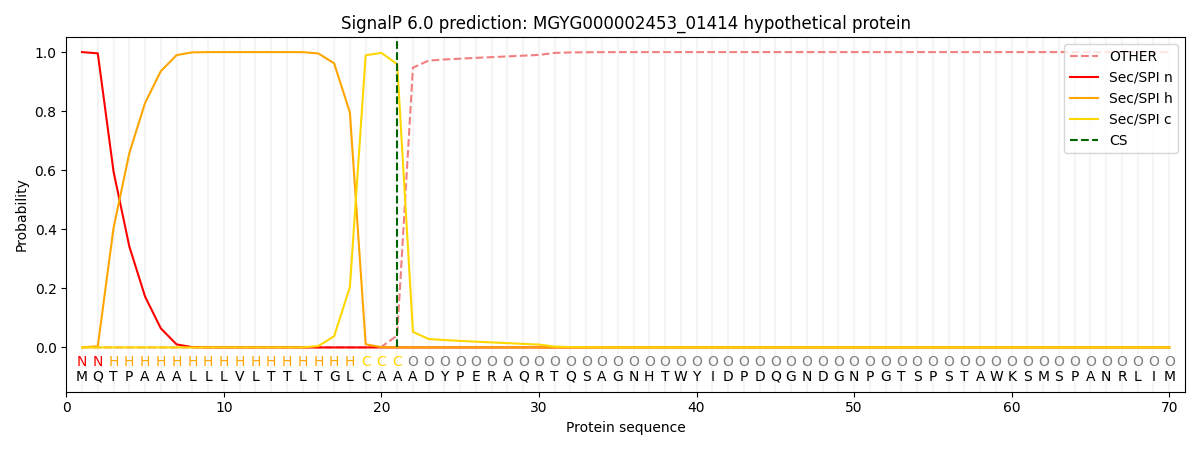
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000250 | 0.999142 | 0.000147 | 0.000182 | 0.000149 | 0.000135 |
