You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002453_01470
You are here: Home > Sequence: MGYG000002453_01470
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Akkermansia muciniphila_B | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Verrucomicrobiales; Akkermansiaceae; Akkermansia; Akkermansia muciniphila_B | |||||||||||
| CAZyme ID | MGYG000002453_01470 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 108567; End: 110480 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 544 | 632 | 6.2e-19 | 0.6791044776119403 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 1.85e-27 | 500 | 636 | 2 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 8.72e-21 | 497 | 636 | 1 | 145 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| pfam12044 | Metallopep | 1.66e-08 | 112 | 306 | 138 | 385 | Putative peptidase family. This family of proteins is functionally uncharacterized. However, it does contain an HEXXH motif characteristic of metallopeptidases. This protein is found mainly in fungi. Proteins in this family are typically between 625 to 773 amino acids in length. |
| cd04275 | ZnMc_pappalysin_like | 0.003 | 235 | 269 | 130 | 166 | Zinc-dependent metalloprotease, pappalysin_like subfamily. The pregnancy-associated plasma protein A (PAPP-A or pappalysin-1) cleaves insulin-like growth factor-binding proteins 4 and 5, thereby promoting cell growth by releasing bound growth factor. This model includes pappalysins and related metalloprotease domains from all three kingdoms of life. The three-dimensional structure of an archaeal representative, ulilysin, has been solved. |
| pfam10462 | Peptidase_M66 | 0.006 | 247 | 296 | 198 | 250 | Peptidase M66. This family of metallopeptidases contains StcE, a virulence factor found in Shiga toxigenic Escherichia coli organisms. StcE peptidase cleaves C1 esterase inhibitor. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHV75710.1 | 0.0 | 1 | 637 | 1 | 637 |
| QHV63340.1 | 0.0 | 1 | 637 | 1 | 637 |
| QUY59663.1 | 0.0 | 1 | 637 | 1 | 637 |
| QWP47761.1 | 0.0 | 4 | 637 | 1 | 634 |
| QWP72337.1 | 0.0 | 4 | 637 | 1 | 634 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Z2D_A | 4.62e-15 | 79 | 396 | 16 | 321 | ChainA, O-glycan protease [Akkermansia muciniphila ATCC BAA-835] |
| 6Z2O_A | 4.65e-15 | 79 | 396 | 17 | 322 | ChainA, O-glycan protease [Akkermansia muciniphila ATCC BAA-835],6Z2Q_A Chain A, O-glycan protease [Akkermansia muciniphila ATCC BAA-835] |
| 6Z2P_A | 4.75e-13 | 79 | 396 | 17 | 322 | ChainA, O-glycan protease [Akkermansia muciniphila ATCC BAA-835] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
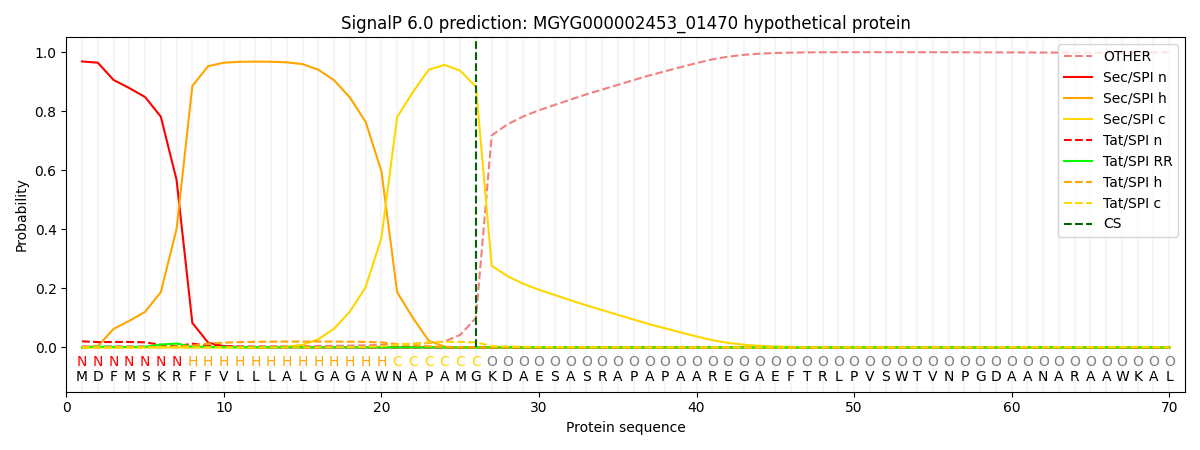
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.006655 | 0.961459 | 0.007701 | 0.023079 | 0.000764 | 0.000302 |
