You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002455_00257
You are here: Home > Sequence: MGYG000002455_00257
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides cellulosilyticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides cellulosilyticus | |||||||||||
| CAZyme ID | MGYG000002455_00257 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 326588; End: 329431 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14670 | BslA_like | 1.75e-33 | 122 | 259 | 2 | 128 | Bacterial immunoglobulin-like hydrophobin BslA and similar proteins. BslA (YuaB) is a protein from Bacillus subtilis acting as a hydrophobin, which forms surface layers around biofilms and participates in biofilm assembly. BslA contains an unusually hydrophobic "cap structure", which is essential for its activity and for the ability of bacteria to form hydrophobic, non-wetting biofilms. A number of domains in various proteins from Bacilli and other bacterial lineages appear related to BslA, but do not conserve the hydrophobic cap. |
| COG5434 | Pgu1 | 1.66e-18 | 377 | 778 | 84 | 448 | Polygalacturonase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT90412.1 | 0.0 | 1 | 947 | 1 | 947 |
| ALJ58475.1 | 0.0 | 1 | 947 | 1 | 947 |
| QDO71142.1 | 0.0 | 1 | 946 | 1 | 946 |
| AWI10257.1 | 0.0 | 39 | 946 | 31 | 949 |
| ACU05945.1 | 0.0 | 38 | 946 | 44 | 965 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
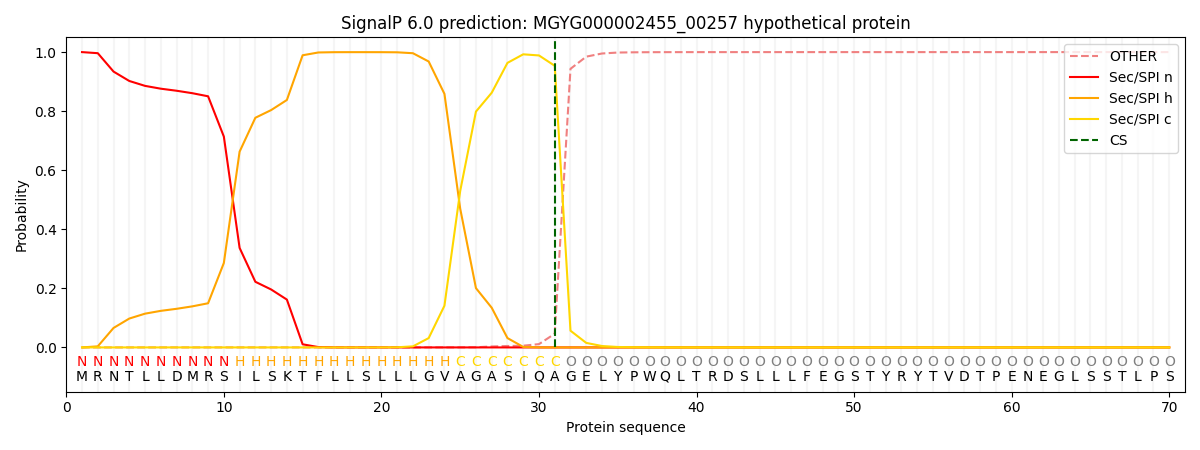
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000415 | 0.998912 | 0.000210 | 0.000159 | 0.000149 | 0.000141 |
