You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002459_01022
You are here: Home > Sequence: MGYG000002459_01022
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
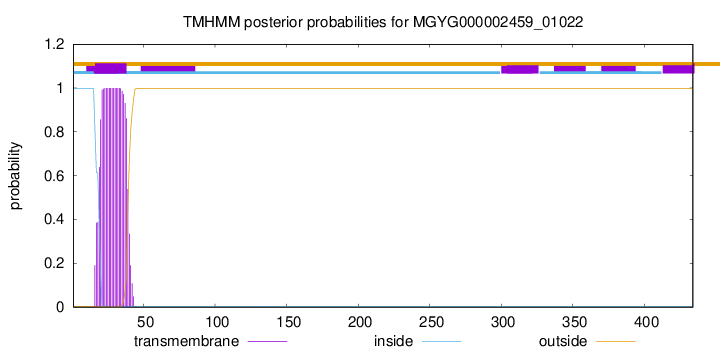
TMHMM annotations
Basic Information help
| Species | Bifidobacterium animalis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Bifidobacterium; Bifidobacterium animalis | |||||||||||
| CAZyme ID | MGYG000002459_01022 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1174540; End: 1175844 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 159 | 389 | 1.3e-37 | 0.9768518518518519 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 2.50e-49 | 99 | 433 | 1 | 318 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 1.56e-34 | 100 | 426 | 1 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| PRK05337 | PRK05337 | 1.18e-16 | 164 | 366 | 55 | 256 | beta-hexosaminidase; Provisional |
| COG3147 | DedD | 0.009 | 6 | 167 | 43 | 222 | Cell division protein DedD (periplasmic protein involved in septation) [Cell cycle control, cell division, chromosome partitioning]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QYU55325.1 | 1.50e-301 | 1 | 434 | 46 | 479 |
| AIA33024.1 | 1.50e-301 | 1 | 434 | 46 | 479 |
| AFJ18331.1 | 1.50e-301 | 1 | 434 | 46 | 479 |
| QRI03329.1 | 1.50e-301 | 1 | 434 | 46 | 479 |
| ACS46383.1 | 1.50e-301 | 1 | 434 | 46 | 479 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5BU9_A | 4.60e-68 | 99 | 427 | 5 | 337 | Crystalstructure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 [Beutenbergia cavernae DSM 12333],5BU9_B Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 [Beutenbergia cavernae DSM 12333] |
| 6K5J_A | 3.57e-22 | 97 | 427 | 9 | 337 | Structureof a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 [Paenibacillaceae] |
| 4ZM6_A | 4.52e-20 | 100 | 390 | 8 | 299 | Aunique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase [Rhizomucor miehei CAU432],4ZM6_B A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase [Rhizomucor miehei CAU432] |
| 6GFV_A | 2.14e-19 | 99 | 427 | 16 | 337 | Mtuberculosis LpqI [Mycobacterium tuberculosis H37Rv],6GFV_B M tuberculosis LpqI [Mycobacterium tuberculosis H37Rv] |
| 7VI6_A | 3.16e-17 | 123 | 405 | 18 | 290 | ChainA, Beta-N-acetylhexosaminidase [Akkermansia muciniphila ATCC BAA-835],7VI6_B Chain B, Beta-N-acetylhexosaminidase [Akkermansia muciniphila ATCC BAA-835],7VI7_A Chain A, Beta-N-acetylhexosaminidase [Akkermansia muciniphila ATCC BAA-835],7VI7_B Chain B, Beta-N-acetylhexosaminidase [Akkermansia muciniphila ATCC BAA-835] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D4AQ52 | 6.31e-22 | 98 | 396 | 18 | 315 | Uncharacterized secreted glycosyl hydrolase ARB_06359 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_06359 PE=1 SV=1 |
| L7N6B0 | 1.85e-18 | 99 | 427 | 59 | 380 | Beta-hexosaminidase LpqI OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=lpqI PE=1 SV=1 |
| A0A0H3M1P5 | 4.53e-18 | 99 | 427 | 59 | 380 | Beta-hexosaminidase LpqI OS=Mycobacterium bovis (strain BCG / Pasteur 1173P2) OX=410289 GN=lpqI PE=3 SV=1 |
| Q0HVQ8 | 3.22e-17 | 125 | 386 | 20 | 276 | Beta-hexosaminidase OS=Shewanella sp. (strain MR-7) OX=60481 GN=nagZ PE=3 SV=1 |
| A0KXK3 | 5.87e-17 | 125 | 386 | 20 | 276 | Beta-hexosaminidase OS=Shewanella sp. (strain ANA-3) OX=94122 GN=nagZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
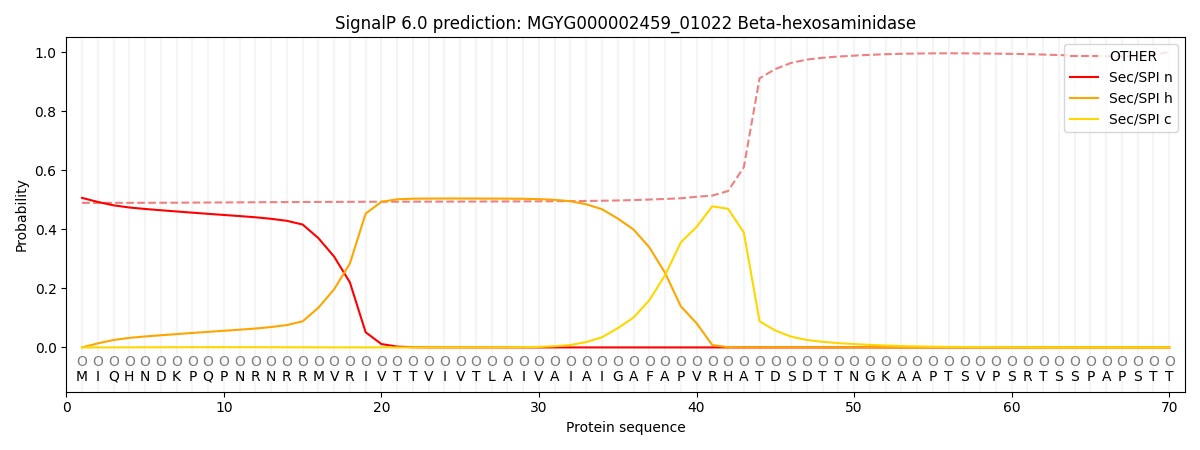
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.509870 | 0.475668 | 0.005296 | 0.004203 | 0.001671 | 0.003297 |