You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002463_03842
You are here: Home > Sequence: MGYG000002463_03842
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
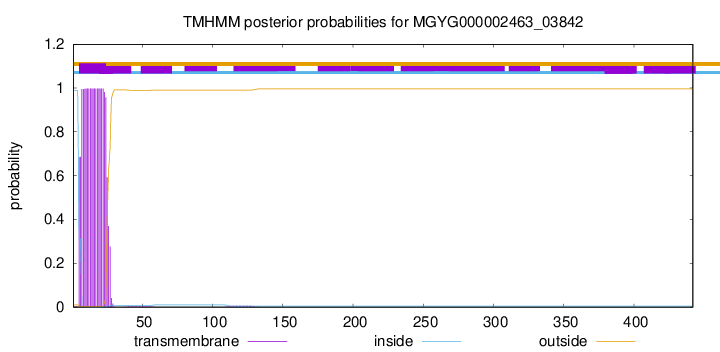
TMHMM annotations
Basic Information help
| Species | Pseudomonas aeruginosa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas; Pseudomonas aeruginosa | |||||||||||
| CAZyme ID | MGYG000002463_03842 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10134; End: 11462 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 88 | 394 | 1.5e-37 | 0.7238979118329466 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3664 | XynB | 2.76e-167 | 23 | 442 | 8 | 424 | Beta-xylosidase [Carbohydrate transport and metabolism]. |
| pfam01229 | Glyco_hydro_39 | 4.29e-08 | 136 | 396 | 120 | 384 | Glycosyl hydrolases family 39. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGY69805.1 | 0.0 | 1 | 442 | 1 | 442 |
| QDY00834.1 | 0.0 | 1 | 442 | 1 | 442 |
| ARI98072.1 | 0.0 | 1 | 442 | 1 | 442 |
| ASD03991.1 | 0.0 | 1 | 442 | 1 | 442 |
| AOX27121.1 | 0.0 | 1 | 442 | 1 | 442 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4ZN2_A | 1.05e-314 | 30 | 442 | 4 | 416 | Glycosylhydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_B Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_C Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_D Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
| 5BX9_A | 1.49e-314 | 31 | 442 | 5 | 416 | Structureof PslG from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5BXA_A Structure of PslG from Pseudomonas aeruginosa in complex with mannose [Pseudomonas aeruginosa PAO1] |
| 4EKJ_A | 2.15e-06 | 68 | 369 | 51 | 372 | ChainA, Beta-xylosidase [Caulobacter vibrioides] |
| 4M29_A | 3.76e-06 | 68 | 369 | 51 | 372 | Structureof a GH39 Beta-xylosidase from Caulobacter crescentus [Caulobacter vibrioides CB15] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
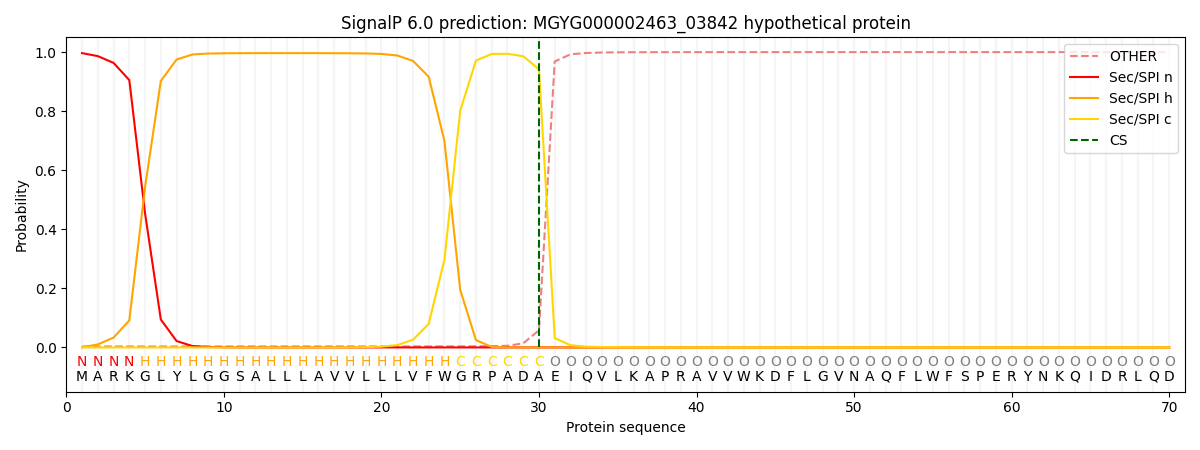
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.006475 | 0.992118 | 0.000606 | 0.000344 | 0.000217 | 0.000215 |