You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002463_04181
You are here: Home > Sequence: MGYG000002463_04181
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pseudomonas aeruginosa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas; Pseudomonas aeruginosa | |||||||||||
| CAZyme ID | MGYG000002463_04181 | |||||||||||
| CAZy Family | CBM22 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 41217; End: 43016 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM22 | 458 | 580 | 1.3e-20 | 0.9312977099236641 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13582 | Reprolysin_3 | 4.75e-35 | 46 | 175 | 1 | 122 | Metallo-peptidase family M12B Reprolysin-like. This zinc-binding metallo-peptidase has the characteristic binding motif HExxGHxxGxxH of Reprolysin-like peptidases of family M12B. |
| pfam13688 | Reprolysin_5 | 2.30e-14 | 22 | 213 | 2 | 181 | Metallo-peptidase family M12. |
| pfam13583 | Reprolysin_4 | 5.34e-13 | 22 | 242 | 2 | 198 | Metallo-peptidase family M12B Reprolysin-like. This zinc-binding metallo-peptidase has the characteristic binding motif HExxGHxxGxxH of Reprolysin-like peptidases of family M12B. |
| NF033511 | metallo_CpaA | 2.01e-10 | 24 | 264 | 379 | 575 | metalloendopeptidase CpaA. |
| pfam02018 | CBM_4_9 | 5.56e-06 | 458 | 584 | 7 | 132 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANA72715.1 | 0.0 | 1 | 599 | 1 | 599 |
| QKE77829.1 | 0.0 | 1 | 599 | 1 | 599 |
| QKQ60417.1 | 0.0 | 1 | 599 | 1 | 599 |
| AID85426.1 | 0.0 | 1 | 599 | 1 | 599 |
| AVK17941.1 | 0.0 | 1 | 599 | 1 | 599 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9I060 | 0.0 | 1 | 599 | 1 | 599 | Peptidyl-Asp metalloendopeptidase OS=Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) OX=208964 GN=mep72 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
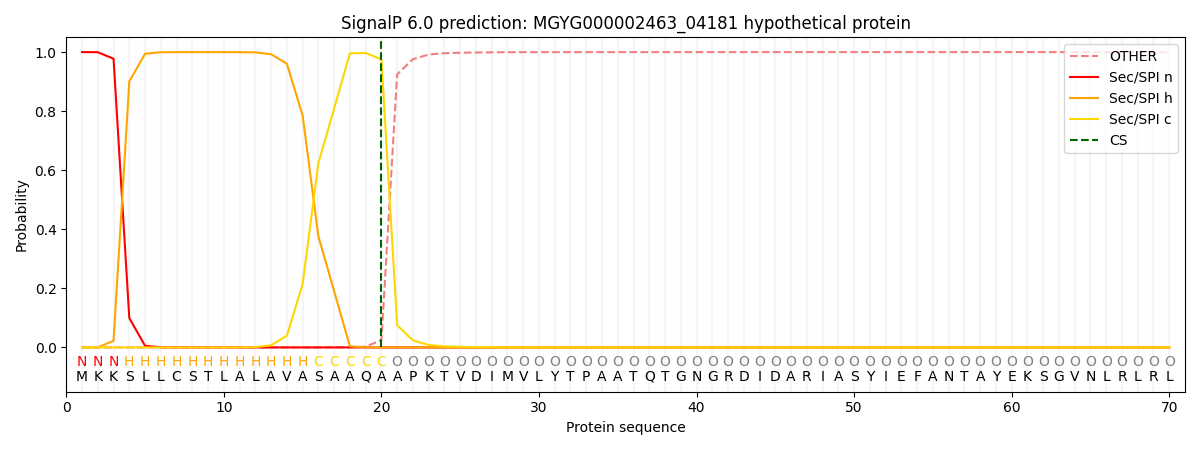
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001090 | 0.997281 | 0.000366 | 0.000442 | 0.000416 | 0.000366 |
