You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002466_01410
You are here: Home > Sequence: MGYG000002466_01410
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Yersinia frederiksenii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Yersinia; Yersinia frederiksenii | |||||||||||
| CAZyme ID | MGYG000002466_01410 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 175123; End: 175602 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 28 | 145 | 2.5e-21 | 0.9274193548387096 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 2.54e-17 | 26 | 145 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 1.91e-07 | 27 | 129 | 14 | 126 | Substituted updates: Jan 31, 2002 |
| pfam17132 | Glyco_hydro_106 | 1.06e-05 | 26 | 129 | 175 | 282 | alpha-L-rhamnosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ATM94021.1 | 3.82e-110 | 1 | 159 | 1 | 159 |
| QAX80876.1 | 4.12e-105 | 1 | 159 | 1 | 159 |
| AHM71405.2 | 1.68e-104 | 1 | 159 | 1 | 159 |
| VTP84873.1 | 3.25e-102 | 1 | 159 | 1 | 159 |
| AJI82176.1 | 3.25e-102 | 1 | 159 | 1 | 159 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2JD9_A | 7.82e-90 | 20 | 159 | 6 | 145 | Structureof a pectin binding carbohydrate binding module determined in an orthorhombic crystal form. [Yersinia enterocolitica],2JDA_A Structure of a pectin binding carbohydrate binding module determined in an monoclinic crystal form. [Yersinia enterocolitica],2JDA_B Structure of a pectin binding carbohydrate binding module determined in an monoclinic crystal form. [Yersinia enterocolitica] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
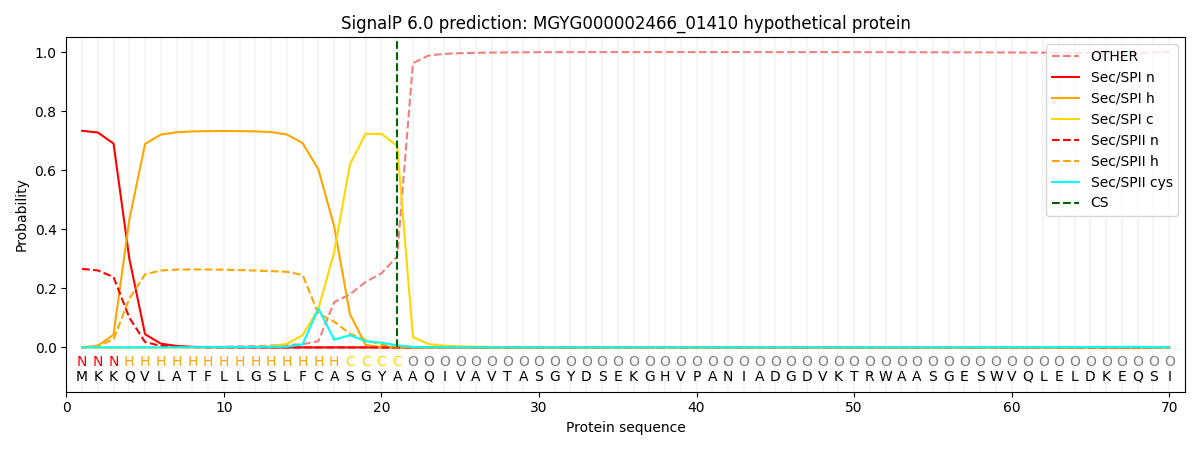
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001879 | 0.721922 | 0.275100 | 0.000515 | 0.000300 | 0.000270 |

