You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002473_00579
You are here: Home > Sequence: MGYG000002473_00579
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Victivallis vadensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; Victivallis vadensis | |||||||||||
| CAZyme ID | MGYG000002473_00579 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3249; End: 5666 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 382 | 752 | 2.7e-49 | 0.8723897911832946 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01229 | Glyco_hydro_39 | 7.53e-37 | 327 | 799 | 3 | 487 | Glycosyl hydrolases family 39. |
| COG3664 | XynB | 1.49e-09 | 364 | 771 | 21 | 397 | Beta-xylosidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM45825.1 | 0.0 | 13 | 803 | 16 | 804 |
| AVM44872.1 | 3.31e-162 | 10 | 799 | 120 | 923 |
| QPC92966.1 | 2.41e-29 | 392 | 787 | 71 | 463 |
| AXC12885.1 | 3.22e-29 | 322 | 768 | 26 | 473 |
| QPC95981.1 | 2.54e-28 | 392 | 787 | 71 | 463 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4M29_A | 3.12e-20 | 391 | 801 | 76 | 498 | Structureof a GH39 Beta-xylosidase from Caulobacter crescentus [Caulobacter vibrioides CB15] |
| 4EKJ_A | 5.51e-20 | 391 | 801 | 76 | 498 | ChainA, Beta-xylosidase [Caulobacter vibrioides] |
| 1PX8_A | 3.60e-16 | 397 | 667 | 78 | 357 | Crystalstructure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase [Thermoanaerobacterium saccharolyticum],1PX8_B Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase [Thermoanaerobacterium saccharolyticum],1UHV_A Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase [Thermoanaerobacterium saccharolyticum],1UHV_B Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase [Thermoanaerobacterium saccharolyticum],1UHV_C Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase [Thermoanaerobacterium saccharolyticum],1UHV_D Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase [Thermoanaerobacterium saccharolyticum] |
| 6YYH_A | 6.91e-16 | 397 | 667 | 101 | 380 | Crystalstructure of beta-D-xylosidase from Dictyoglomus thermophilum in ligand-free form [Dictyoglomus thermophilum H-6-12],6YYH_B Crystal structure of beta-D-xylosidase from Dictyoglomus thermophilum in ligand-free form [Dictyoglomus thermophilum H-6-12],6YYI_A Crystal structure of beta-D-xylosidase from Dictyoglomus thermophilum bound to beta-D-xylopyranose [Dictyoglomus thermophilum H-6-12],6YYI_B Crystal structure of beta-D-xylosidase from Dictyoglomus thermophilum bound to beta-D-xylopyranose [Dictyoglomus thermophilum H-6-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23552 | 3.36e-28 | 391 | 799 | 75 | 485 | Beta-xylosidase OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=xynB PE=3 SV=1 |
| O30360 | 1.17e-16 | 397 | 667 | 78 | 357 | Beta-xylosidase OS=Thermoanaerobacterium saccharolyticum (strain DSM 8691 / JW/SL-YS485) OX=1094508 GN=xynB PE=3 SV=1 |
| P36906 | 2.48e-14 | 397 | 667 | 78 | 357 | Beta-xylosidase OS=Thermoanaerobacterium saccharolyticum OX=28896 GN=xynB PE=1 SV=1 |
| Q01634 | 2.45e-10 | 318 | 803 | 22 | 544 | Alpha-L-iduronidase OS=Canis lupus familiaris OX=9615 GN=IDUA PE=1 SV=1 |
| P48441 | 4.95e-09 | 327 | 773 | 22 | 501 | Alpha-L-iduronidase OS=Mus musculus OX=10090 GN=Idua PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
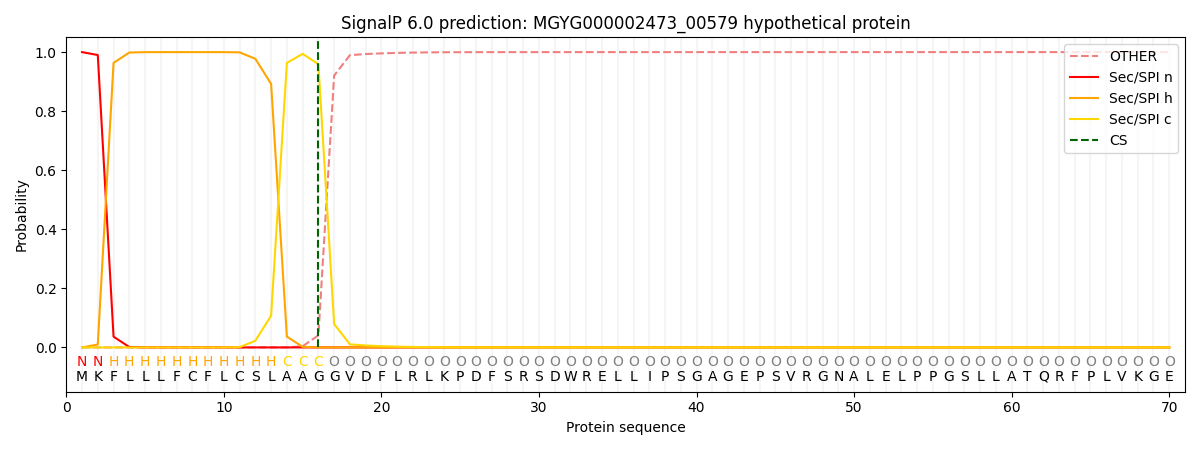
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000433 | 0.998907 | 0.000172 | 0.000159 | 0.000149 | 0.000151 |
