You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002473_01523
You are here: Home > Sequence: MGYG000002473_01523
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Victivallis vadensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; Victivallis vadensis | |||||||||||
| CAZyme ID | MGYG000002473_01523 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 22509; End: 24377 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 99 | 257 | 9.6e-24 | 0.4385150812064965 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3693 | XynA | 6.12e-06 | 58 | 172 | 67 | 191 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| COG1874 | GanA | 1.21e-04 | 26 | 235 | 19 | 254 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| pfam01301 | Glyco_hydro_35 | 3.22e-04 | 30 | 105 | 17 | 86 | Glycosyl hydrolases family 35. |
| pfam02449 | Glyco_hydro_42 | 0.002 | 36 | 90 | 4 | 64 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44885.1 | 1.38e-120 | 10 | 608 | 6 | 607 |
| QDU57372.1 | 2.27e-83 | 22 | 612 | 27 | 625 |
| AEF80792.1 | 4.32e-29 | 36 | 241 | 39 | 246 |
| QPG71004.1 | 3.82e-28 | 34 | 241 | 57 | 286 |
| BBY81341.1 | 7.04e-27 | 19 | 258 | 40 | 299 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4ZN2_A | 1.30e-10 | 26 | 177 | 22 | 182 | Glycosylhydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_B Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_C Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_D Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
| 5BX9_A | 1.30e-10 | 26 | 177 | 22 | 182 | Structureof PslG from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5BXA_A Structure of PslG from Pseudomonas aeruginosa in complex with mannose [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
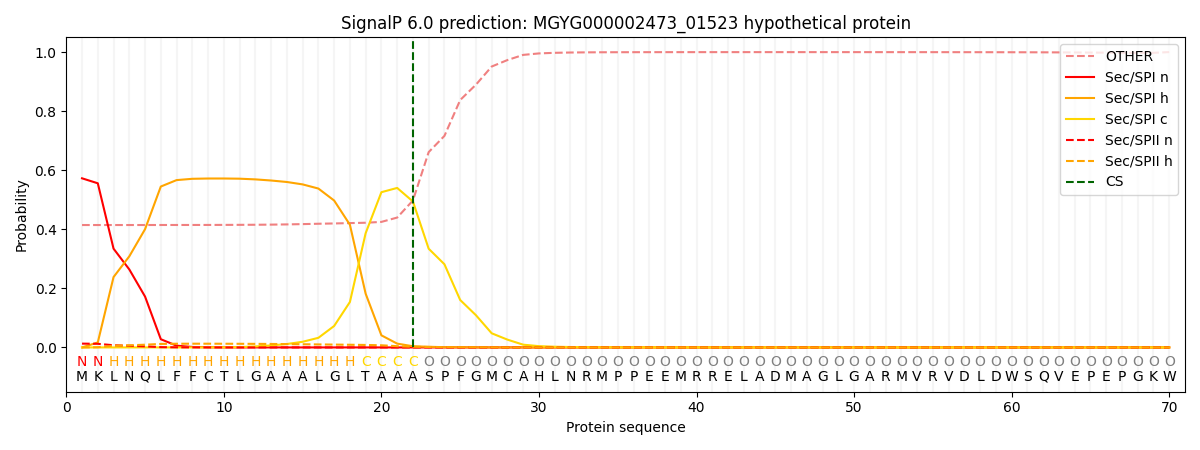
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.432051 | 0.551617 | 0.014890 | 0.000630 | 0.000348 | 0.000457 |

