You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002473_02301
You are here: Home > Sequence: MGYG000002473_02301
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Victivallis vadensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; Victivallis vadensis | |||||||||||
| CAZyme ID | MGYG000002473_02301 | |||||||||||
| CAZy Family | GH87 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4177; End: 5826 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH87 | 21 | 381 | 3.6e-19 | 0.507537688442211 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 6.03e-17 | 14 | 310 | 76 | 397 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam12708 | Pectate_lyase_3 | 1.55e-07 | 22 | 64 | 3 | 46 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| pfam13229 | Beta_helix | 5.99e-04 | 164 | 318 | 1 | 145 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| PLN02188 | PLN02188 | 0.001 | 22 | 65 | 38 | 82 | polygalacturonase/glycoside hydrolase family protein |
| PLN02155 | PLN02155 | 0.003 | 21 | 213 | 28 | 221 | polygalacturonase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM46992.1 | 3.39e-210 | 21 | 536 | 23 | 538 |
| AVM46991.1 | 3.49e-105 | 21 | 431 | 25 | 453 |
| AKK29103.1 | 2.28e-80 | 21 | 386 | 19 | 383 |
| BBY49293.1 | 2.62e-78 | 10 | 367 | 10 | 394 |
| BBX17974.1 | 1.39e-73 | 21 | 368 | 33 | 397 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q44492 | 7.29e-06 | 22 | 165 | 4 | 148 | Mannuronan C5-epimerase AlgE5 OS=Azotobacter vinelandii OX=354 GN=algE5 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
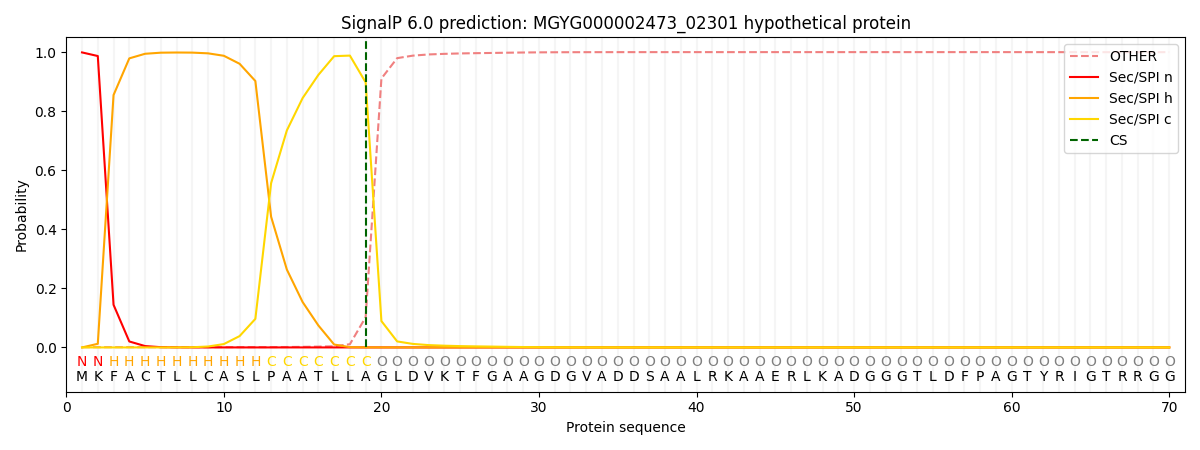
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002259 | 0.996159 | 0.000799 | 0.000277 | 0.000237 | 0.000244 |
