You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002474_03257
You are here: Home > Sequence: MGYG000002474_03257
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
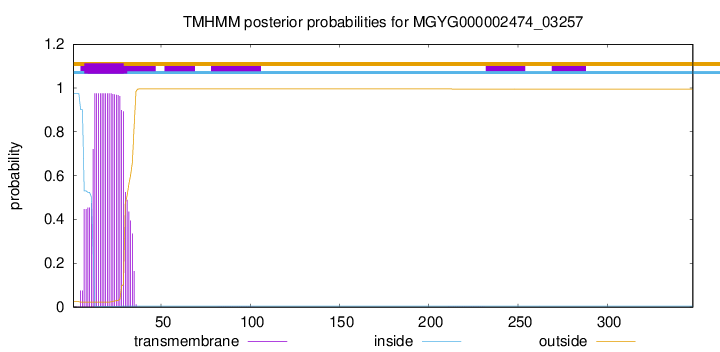
TMHMM annotations
Basic Information help
| Species | Rahnella variigena | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Rahnella; Rahnella variigena | |||||||||||
| CAZyme ID | MGYG000002474_03257 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 53258; End: 54304 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 73 | 338 | 4.4e-25 | 0.6380510440835266 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.05e-11 | 43 | 290 | 29 | 259 | Cellulase (glycosyl hydrolase family 5). |
| COG2723 | BglB | 5.79e-05 | 47 | 177 | 68 | 210 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| PRK09589 | celA | 1.68e-04 | 43 | 143 | 69 | 176 | 6-phospho-beta-glucosidase; Reviewed |
| pfam00232 | Glyco_hydro_1 | 4.15e-04 | 44 | 176 | 64 | 206 | Glycosyl hydrolase family 1. |
| pfam01229 | Glyco_hydro_39 | 8.91e-04 | 105 | 266 | 113 | 270 | Glycosyl hydrolases family 39. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QBJ08916.1 | 5.24e-233 | 1 | 344 | 1 | 344 |
| ATR01912.1 | 7.62e-133 | 27 | 335 | 24 | 332 |
| ATR07087.1 | 7.62e-133 | 27 | 335 | 24 | 332 |
| ATR12484.1 | 7.62e-133 | 27 | 335 | 24 | 332 |
| ATR18209.1 | 7.62e-133 | 27 | 335 | 24 | 332 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4ZN2_A | 2.94e-08 | 44 | 342 | 39 | 329 | Glycosylhydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_B Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_C Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_D Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
| 5BX9_A | 2.94e-08 | 44 | 342 | 39 | 329 | Structureof PslG from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5BXA_A Structure of PslG from Pseudomonas aeruginosa in complex with mannose [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
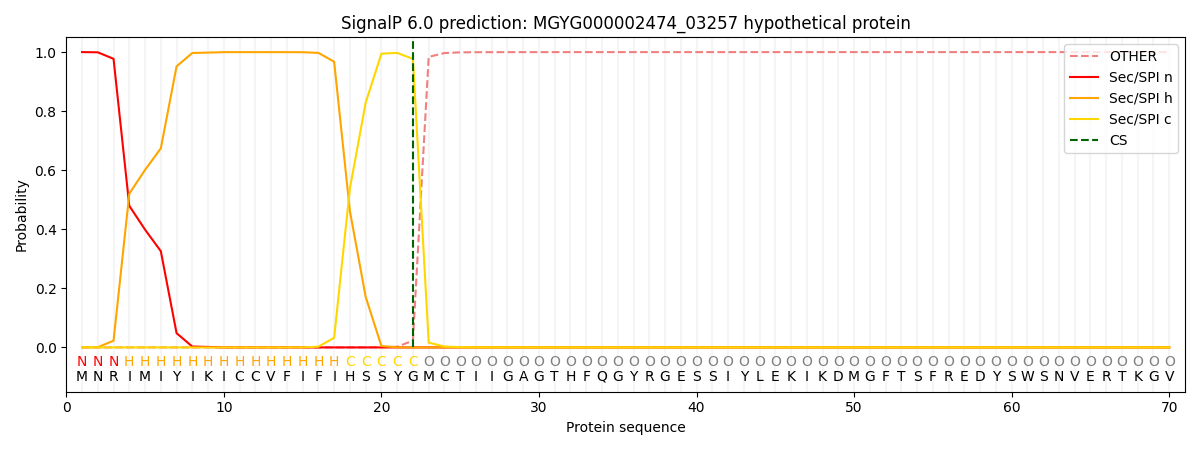
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000460 | 0.998948 | 0.000176 | 0.000139 | 0.000133 | 0.000137 |