You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002476_00227
You are here: Home > Sequence: MGYG000002476_00227
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Yersinia pestis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Yersinia; Yersinia pestis | |||||||||||
| CAZyme ID | MGYG000002476_00227 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 140976; End: 141527 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK13888 | PRK13888 | 6.21e-47 | 45 | 181 | 15 | 148 | conjugal transfer protein TrbN; Provisional |
| cd13400 | LT_IagB-like | 6.56e-28 | 56 | 162 | 2 | 104 | Escherichia coli invasion protein IagB and similar proteins. Lytic transglycosylase-like protein, similar to Escherichia coli invasion protein IagB. IagB is encoded within a pathogenicity island in Salmonella enterica and has been shown to degrade polymeric peptidoglycan. IagB-like invasion proteins are implicated in the invasion of eukaryotic host cells by bacteria. Lytic transglycosylase (LT) catalyzes the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Members of this family resemble the soluble and insoluble membrane-bound LTs in bacteria and the LTs in bacteriophage lambda. |
| PRK12712 | flgJ | 1.46e-04 | 51 | 96 | 211 | 257 | flagellar rod assembly protein/muramidase FlgJ; Provisional |
| cd16899 | LYZ_C_invert | 3.10e-04 | 53 | 96 | 13 | 61 | C-type invertebrate lysozyme. C-type lysozyme proteins of invertebrates, including digestive lysozymes 1 and 2 from Musca domestica, which aid in the use of bacteria as a food source. These lysozymes have high expression in the gut and optimal lytic activity at a lower pH. Other lysozymes in this subfamily have immunological roles. e.g. Anopheles gambiae has eight lysozymes, most of which seem to have immunological roles, those some may function as digestive enzymes in larvae. C-type lysozyme (chicken or conventional type; 1, 4-beta-N-acetylmuramidase) has bacteriolytic properties through the hydolysis of beta-1,4, glyocosidic linkages between N-acetylmuramic acid and N-acetyl-D-glucosamine residues in a peptidoglycan, as well as between N-acetyl-D-glucosamine residues in chitodextrins. |
| pfam00062 | Lys | 5.48e-04 | 65 | 101 | 31 | 68 | C-type lysozyme/alpha-lactalbumin family. Alpha-lactalbumin is the regulatory subunit of lactose synthase, changing the substrate specificity of galactosyltransferase from N-acetylglucosamine to glucose. C-type lysozymes are secreted bacteriolytic enzymes that cleave the peptidoglycan of bacterial cell walls. Structure is a multi-domain, mixed alpha and beta fold, containing four conserved disulfide bonds. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABS45748.1 | 9.41e-115 | 25 | 183 | 1 | 159 |
| QTP13337.1 | 2.95e-74 | 36 | 183 | 27 | 178 |
| QHB30063.1 | 1.97e-64 | 36 | 175 | 24 | 164 |
| BBU46966.1 | 1.97e-64 | 36 | 175 | 24 | 164 |
| AOX11182.1 | 5.62e-64 | 36 | 175 | 24 | 164 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
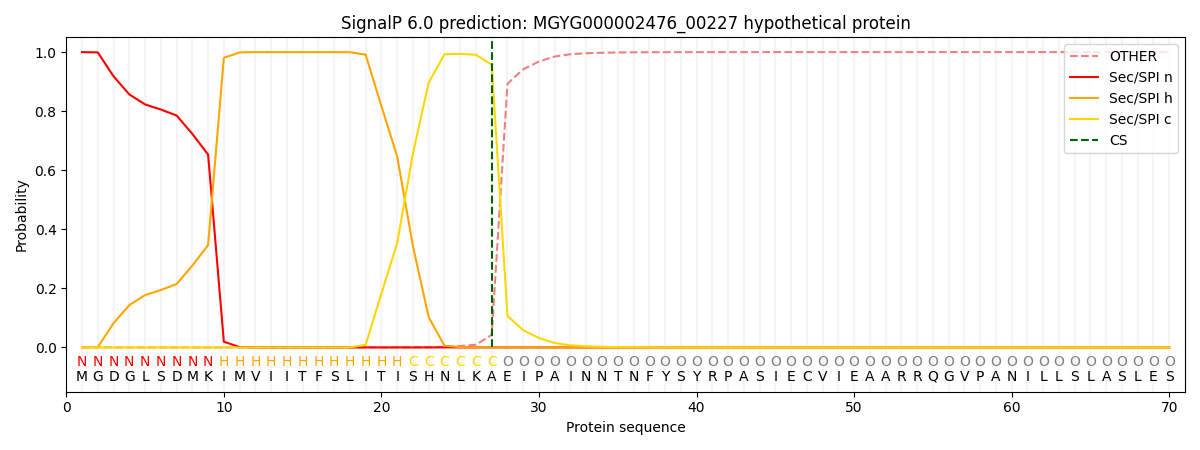
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000214 | 0.999185 | 0.000162 | 0.000140 | 0.000136 | 0.000127 |
