You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002478_00680
You are here: Home > Sequence: MGYG000002478_00680
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola dorei | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola dorei | |||||||||||
| CAZyme ID | MGYG000002478_00680 | |||||||||||
| CAZy Family | GH136 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 961782; End: 963296 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH136 | 24 | 466 | 8.6e-91 | 0.8716904276985743 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 7.47e-04 | 148 | 288 | 8 | 138 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 0.003 | 173 | 334 | 1 | 133 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT85993.1 | 0.0 | 1 | 504 | 1 | 504 |
| ALA72617.1 | 0.0 | 1 | 504 | 1 | 504 |
| QJR78357.1 | 0.0 | 1 | 504 | 1 | 504 |
| QJR69871.1 | 0.0 | 1 | 504 | 1 | 504 |
| QJR74203.1 | 0.0 | 1 | 504 | 1 | 504 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6KQT_A | 2.03e-28 | 23 | 349 | 246 | 617 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein [Eubacterium ramulus ATCC 29099] |
| 6KQS_A | 2.11e-27 | 23 | 349 | 246 | 617 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative [Eubacterium ramulus ATCC 29099] |
| 7V6M_A | 2.38e-25 | 24 | 416 | 11 | 482 | ChainA, Fibronectin type III domain-containing protein [Tyzzerella nexilis] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
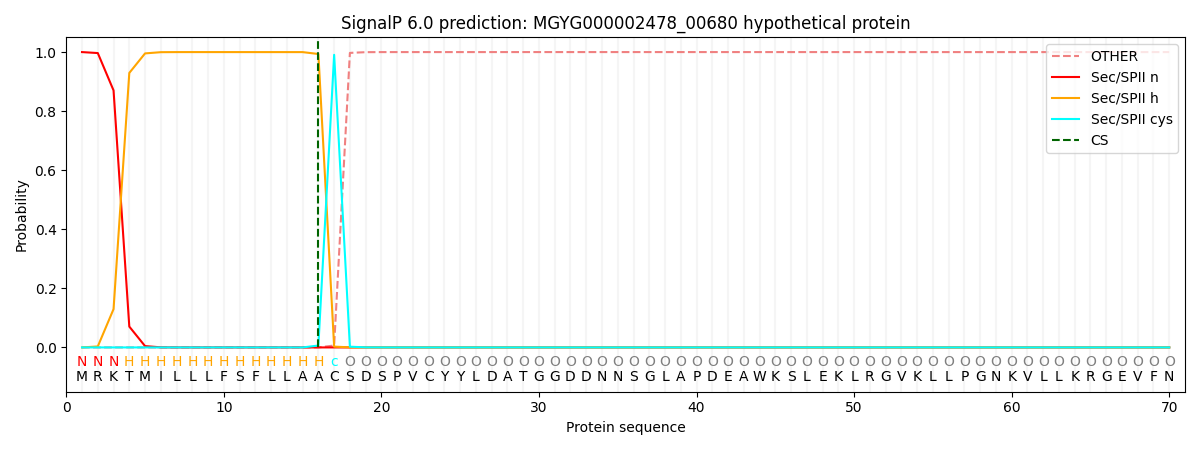
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000003 | 1.000056 | 0.000000 | 0.000000 | 0.000000 |
