You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002478_04021
You are here: Home > Sequence: MGYG000002478_04021
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola dorei | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola dorei | |||||||||||
| CAZyme ID | MGYG000002478_04021 | |||||||||||
| CAZy Family | GH95 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4801050; End: 4803566 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH95 | 43 | 754 | 2.3e-61 | 0.9792243767313019 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1554 | ATH1 | 2.88e-05 | 223 | 537 | 207 | 549 | Trehalose and maltose hydrolase (possible phosphorylase) [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHW32096.1 | 1.13e-40 | 123 | 794 | 441 | 1081 |
| AWS40658.1 | 2.97e-33 | 146 | 794 | 307 | 926 |
| AEI44617.1 | 1.87e-29 | 46 | 795 | 33 | 823 |
| AFC32407.1 | 4.02e-29 | 46 | 795 | 3 | 793 |
| AFH64718.1 | 5.31e-29 | 46 | 795 | 3 | 793 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2RDY_A | 1.22e-10 | 316 | 794 | 329 | 789 | ChainA, BH0842 protein [Halalkalibacterium halodurans C-125],2RDY_B Chain B, BH0842 protein [Halalkalibacterium halodurans C-125] |
| 4UFC_A | 1.29e-08 | 435 | 754 | 448 | 743 | Crystalstructure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus],4UFC_B Crystal structure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus] |
| 7FE3_A | 7.21e-07 | 258 | 483 | 243 | 462 | ChainA, Candidate alpha glycoside phosphorylase Glycoside hydrolase family 65 [Flavobacterium johnsoniae UW101],7FE3_B Chain B, Candidate alpha glycoside phosphorylase Glycoside hydrolase family 65 [Flavobacterium johnsoniae UW101],7FE3_C Chain C, Candidate alpha glycoside phosphorylase Glycoside hydrolase family 65 [Flavobacterium johnsoniae UW101],7FE4_A Chain A, Candidate alpha glycoside phosphorylase Glycoside hydrolase family 65 [Flavobacterium johnsoniae UW101],7FE4_B Chain B, Candidate alpha glycoside phosphorylase Glycoside hydrolase family 65 [Flavobacterium johnsoniae UW101],7FE4_C Chain C, Candidate alpha glycoside phosphorylase Glycoside hydrolase family 65 [Flavobacterium johnsoniae UW101] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
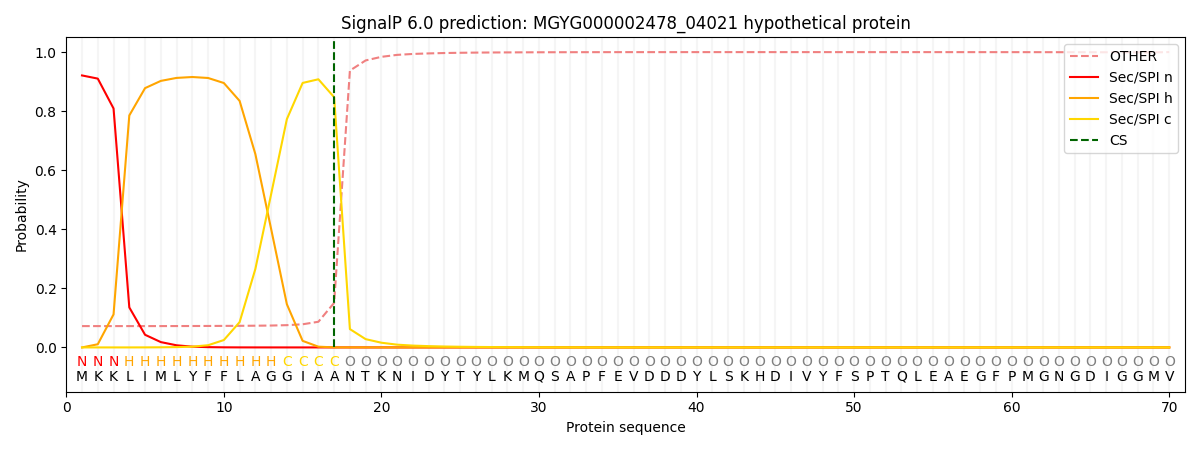
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.080141 | 0.910808 | 0.008007 | 0.000397 | 0.000290 | 0.000305 |
