You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002488_00828
You are here: Home > Sequence: MGYG000002488_00828
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Proteus penneri | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Proteus; Proteus penneri | |||||||||||
| CAZyme ID | MGYG000002488_00828 | |||||||||||
| CAZy Family | PL8 | |||||||||||
| CAZyme Description | Chondroitin sulfate ABC endolyase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 666031; End: 666756 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam09092 | Lyase_N | 9.56e-80 | 38 | 230 | 1 | 166 | Lyase, N terminal. Members of this family are predominantly found in chondroitin ABC lyase I, and adopt a jelly-roll fold topology consisting of a two-layered bent beta-sheet sandwich with one short alpha-helix. The convex beta sheet is composed of five antiparallel strands, whilst the concave beta-sheet contains five antiparallel beta-strands with a loop between two consecutive strands folding back onto the concave surface. This domain is required for binding of the protein to long glycosaminoglycan chains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QPT33224.1 | 2.97e-153 | 1 | 233 | 1 | 233 |
| QKD73830.1 | 1.06e-146 | 1 | 233 | 1 | 233 |
| QKD68656.1 | 1.06e-146 | 1 | 233 | 1 | 233 |
| ATM99761.1 | 1.49e-146 | 1 | 233 | 1 | 233 |
| QHP77349.1 | 2.09e-146 | 1 | 233 | 1 | 233 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1HN0_A | 6.12e-143 | 1 | 233 | 1 | 233 | CRYSTALSTRUCTURE OF CHONDROITIN ABC LYASE I FROM PROTEUS VULGARIS AT 1.9 ANGSTROMS RESOLUTION [Proteus vulgaris] |
| 7EIP_A | 6.12e-143 | 1 | 233 | 1 | 233 | ChainA, Chondroitin sulfate ABC endolyase [Proteus vulgaris],7EIQ_A Chain A, Chondroitin sulfate ABC endolyase [Proteus vulgaris],7EIR_A Chain A, Chondroitin sulfate ABC endolyase [Proteus vulgaris],7EIS_A Chain A, Chondroitin sulfate ABC endolyase [Proteus vulgaris] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P59807 | 3.35e-142 | 1 | 233 | 1 | 233 | Chondroitin sulfate ABC endolyase OS=Proteus vulgaris OX=585 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
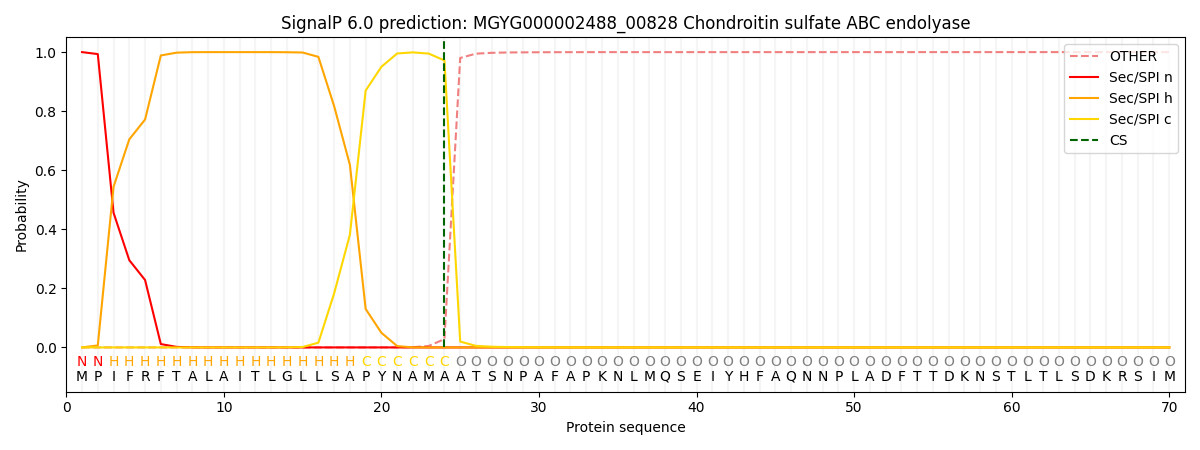
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000289 | 0.998935 | 0.000202 | 0.000193 | 0.000179 | 0.000155 |
