You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002493_01909
Basic Information
help
| Species |
Enterococcus_B pernyi
|
| Lineage |
Bacteria; Firmicutes; Bacilli; Lactobacillales; Enterococcaceae; Enterococcus_B; Enterococcus_B pernyi
|
| CAZyme ID |
MGYG000002493_01909
|
| CAZy Family |
GT2 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000002493 |
3324684 |
Isolate |
Chile |
South America |
|
| Gene Location |
Start: 94242;
End: 94805
Strand: -
|
No EC number prediction in MGYG000002493_01909.
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| PRK11498
|
bcsA |
2.53e-05 |
3 |
59 |
609 |
666 |
cellulose synthase catalytic subunit; Provisional |
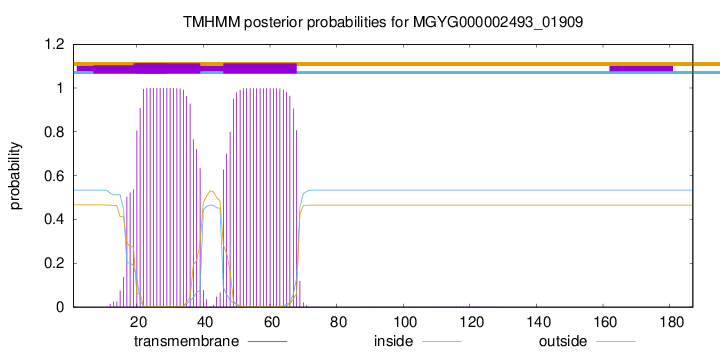
This protein is predicted as OTHER
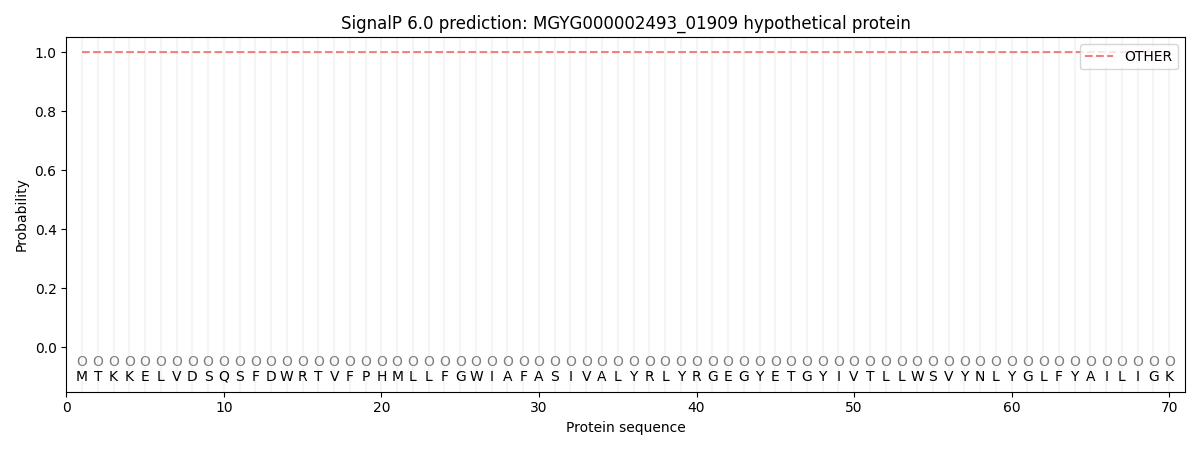
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
1.000033
|
0.000001
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|