You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002493_03002
You are here: Home > Sequence: MGYG000002493_03002
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Enterococcus_B pernyi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Enterococcaceae; Enterococcus_B; Enterococcus_B pernyi | |||||||||||
| CAZyme ID | MGYG000002493_03002 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11555; End: 12664 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 86 | 293 | 1.5e-59 | 0.8366336633663366 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00656 | Amb_all | 5.51e-62 | 87 | 294 | 12 | 190 | Amb_all domain. |
| COG3866 | PelB | 1.63e-59 | 3 | 365 | 5 | 340 | Pectate lyase [Carbohydrate transport and metabolism]. |
| pfam00544 | Pec_lyase_C | 1.57e-42 | 87 | 290 | 30 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCJ57963.1 | 1.64e-261 | 1 | 369 | 1 | 369 |
| AIM25983.1 | 7.56e-164 | 27 | 365 | 32 | 371 |
| BAK22145.1 | 2.00e-163 | 27 | 365 | 10 | 349 |
| BBP07856.1 | 7.14e-162 | 27 | 365 | 32 | 371 |
| BBD17300.1 | 7.14e-162 | 27 | 365 | 32 | 371 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1VBL_A | 1.82e-44 | 44 | 365 | 79 | 413 | Structureof the thermostable pectate lyase PL 47 [Bacillus sp. TS-47] |
| 1PCL_A | 1.09e-41 | 21 | 290 | 1 | 276 | ChainA, PECTATE LYASE E [Dickeya chrysanthemi] |
| 1OOC_A | 3.79e-34 | 33 | 290 | 22 | 287 | ChainA, Pectate lyase A [Dickeya chrysanthemi],1OOC_B Chain B, Pectate lyase A [Dickeya chrysanthemi],1PE9_A Chain A, Pectate lyase A [Dickeya chrysanthemi],1PE9_B Chain B, Pectate lyase A [Dickeya chrysanthemi] |
| 1JRG_A | 7.24e-33 | 33 | 290 | 22 | 287 | ChainA, Pectate lyase [Dickeya chrysanthemi],1JRG_B Chain B, Pectate lyase [Dickeya chrysanthemi],1JTA_A Chain A, pectate lyase A [Dickeya chrysanthemi] |
| 3ZSC_A | 3.50e-32 | 22 | 283 | 10 | 225 | Catalyticfunction and substrate recognition of the pectate lyase from Thermotoga maritima [Thermotoga maritima] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5AVN4 | 1.45e-43 | 20 | 315 | 34 | 284 | Pectate lyase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyA PE=1 SV=1 |
| P0C1A4 | 4.27e-42 | 15 | 290 | 35 | 324 | Pectate lyase E OS=Dickeya chrysanthemi OX=556 GN=pelE PE=3 SV=1 |
| P0C1A5 | 1.16e-41 | 15 | 290 | 35 | 324 | Pectate lyase E OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelE PE=3 SV=2 |
| P04960 | 2.14e-41 | 15 | 290 | 24 | 306 | Pectate lyase E OS=Dickeya chrysanthemi OX=556 GN=pelE PE=1 SV=1 |
| P18209 | 6.71e-40 | 18 | 290 | 35 | 312 | Pectate lyase D OS=Dickeya chrysanthemi OX=556 GN=pelD PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
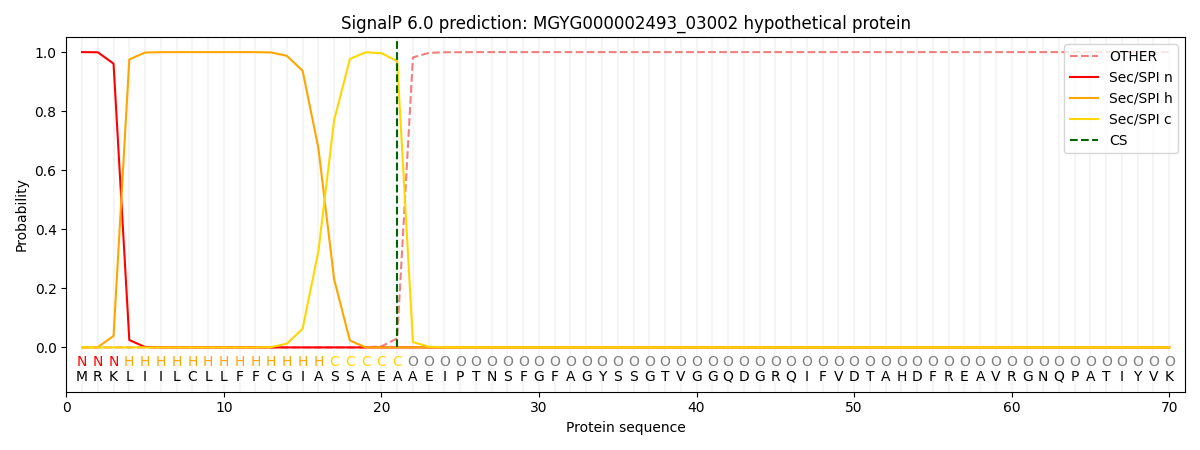
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000272 | 0.998951 | 0.000206 | 0.000193 | 0.000179 | 0.000164 |
