You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002497_04680
You are here: Home > Sequence: MGYG000002497_04680
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
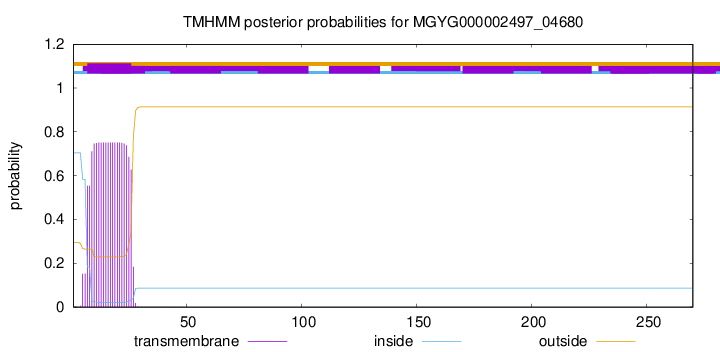
TMHMM annotations
Basic Information help
| Species | Klebsiella aerogenes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Klebsiella; Klebsiella aerogenes | |||||||||||
| CAZyme ID | MGYG000002497_04680 | |||||||||||
| CAZy Family | GH128 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4967491; End: 4968303 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH128 | 51 | 267 | 1.1e-46 | 0.9642857142857143 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam11790 | Glyco_hydro_cc | 1.08e-42 | 49 | 267 | 14 | 235 | Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QEV98098.1 | 2.44e-209 | 1 | 270 | 3 | 272 |
| AXR70442.1 | 2.44e-209 | 1 | 270 | 3 | 272 |
| CZQ24428.1 | 1.79e-206 | 1 | 270 | 1 | 270 |
| BAT23785.1 | 1.11e-205 | 1 | 270 | 3 | 272 |
| AXR70414.1 | 1.37e-127 | 1 | 270 | 1 | 268 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6UAQ_A | 3.39e-13 | 29 | 243 | 25 | 242 | Crystalstructure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) [Amycolatopsis mediterranei],6UAR_A Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaritriose [Amycolatopsis mediterranei] |
| 6UFL_A | 8.66e-13 | 29 | 243 | 25 | 242 | Crystalstructure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199Q mutant) from Amycolatopsis mediterranei (AmGH128_I) in the complex with laminarihexaose [Amycolatopsis mediterranei],6UFZ_A Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199Q mutant) from Amycolatopsis mediterranei (AmGH128_I) [Amycolatopsis mediterranei] |
| 6UAS_A | 2.20e-12 | 29 | 243 | 25 | 242 | Crystalstructure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199A mutant) from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaripentaose [Amycolatopsis mediterranei] |
| 6UAT_A | 2.20e-12 | 29 | 243 | 25 | 242 | Crystalstructure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E102A mutant) from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaripentaose [Amycolatopsis mediterranei],6UAU_A Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E102A mutant) from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaritriose and laminaribiose [Amycolatopsis mediterranei] |
| 6UBC_A | 1.08e-09 | 53 | 267 | 219 | 436 | ChainA, Glyco_hydro_cc domain-containing protein [Cryptococcus neoformans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q09788 | 2.21e-24 | 27 | 267 | 294 | 528 | Alkali-sensitive linkage protein 1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=asl1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
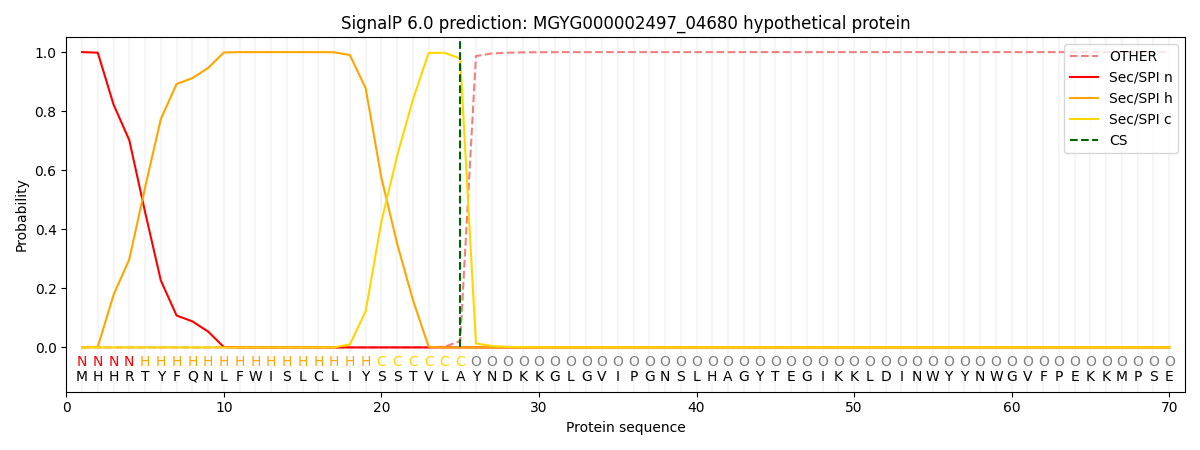
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000248 | 0.999152 | 0.000147 | 0.000155 | 0.000145 | 0.000131 |