You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002520_03228
You are here: Home > Sequence: MGYG000002520_03228
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
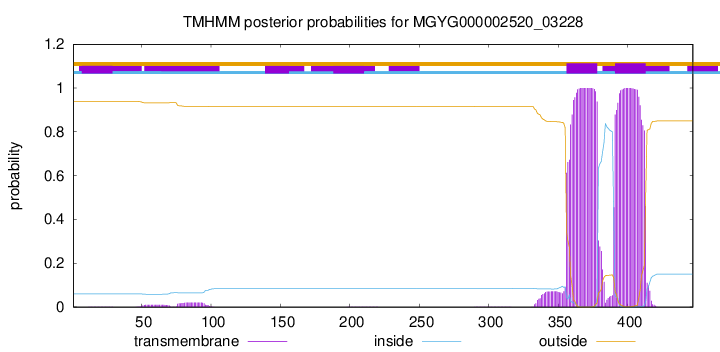
TMHMM annotations
Basic Information help
| Species | Pluralibacter gergoviae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Pluralibacter; Pluralibacter gergoviae | |||||||||||
| CAZyme ID | MGYG000002520_03228 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | Undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3501792; End: 3503135 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 175 | 292 | 5.2e-20 | 0.7294117647058823 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK10714 | PRK10714 | 0.0 | 174 | 447 | 51 | 324 | undecaprenyl phosphate 4-deoxy-4-formamido-L-arabinose transferase; Provisional |
| PRK11658 | PRK11658 | 2.41e-134 | 1 | 173 | 1 | 173 | UDP-4-amino-4-deoxy-L-arabinose aminotransferase. |
| COG0399 | WecE | 1.78e-85 | 1 | 172 | 2 | 173 | dTDP-4-amino-4,6-dideoxygalactose transaminase [Cell wall/membrane/envelope biogenesis]. |
| pfam01041 | DegT_DnrJ_EryC1 | 1.24e-83 | 10 | 173 | 1 | 164 | DegT/DnrJ/EryC1/StrS aminotransferase family. The members of this family are probably all pyridoxal-phosphate-dependent aminotransferase enzymes with a variety of molecular functions. The family includes StsA, StsC and StsS. The aminotransferase activity was demonstrated for purified StsC protein as the L-glutamine:scyllo-inosose aminotransferase EC:2.6.1.50, which catalyzes the first amino transfer in the biosynthesis of the streptidine subunit of streptomycin. |
| cd00616 | AHBA_syn | 3.02e-81 | 16 | 172 | 1 | 157 | 3-amino-5-hydroxybenzoic acid synthase family (AHBA_syn). AHBA_syn family belongs to pyridoxal phosphate (PLP)-dependent aspartate aminotransferase superfamily (fold I). The members of this CD are involved in various biosynthetic pathways for secondary metabolites. Some well studied proteins in this CD are AHBA_synthase, protein product of pleiotropic regulatory gene degT, Arnb aminotransferase and pilin glycosylation protein. The prototype of this family, the AHBA_synthase, is a dimeric PLP dependent enzyme. AHBA_syn is the terminal enzyme of 3-amino-5-hydroxybenzoic acid (AHBA) formation which is involved in the biosynthesis of ansamycin antibiotics, including rifamycin B. Some members of this CD are involved in 4-amino-6-deoxy-monosaccharide D-perosamine synthesis. Perosamine is an important element in the glycosylation of several cell products, such as antibiotics and lipopolysaccharides of gram-positive and gram-negative bacteria. The pilin glycosylation protein encoded by gene pglA, is a galactosyltransferase involved in pilin glycosylation. Additionally, this CD consists of ArnB (PmrH) aminotransferase, a 4-amino-4-deoxy-L-arabinose lipopolysaccharide-modifying enzyme. This CD also consists of several predicted pyridoxal phosphate-dependent enzymes apparently involved in regulation of cell wall biogenesis. The catalytic lysine which is present in all characterized PLP dependent enzymes is replaced by histidine in some members of this CD. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVR04211.1 | 0.0 | 1 | 447 | 1 | 447 |
| AIR01512.1 | 1.29e-189 | 174 | 447 | 53 | 326 |
| AJZ89297.1 | 2.29e-178 | 174 | 447 | 53 | 326 |
| AIR66270.1 | 2.29e-178 | 174 | 447 | 53 | 326 |
| VEB98042.1 | 4.61e-178 | 174 | 447 | 53 | 326 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4OCA_A | 9.31e-94 | 1 | 173 | 7 | 179 | ChainA, UDP-4-amino-4-deoxy-L-arabinose--oxoglutarate aminotransferase [Salmonella enterica subsp. enterica serovar Typhimurium] |
| 1MDX_A | 1.19e-93 | 1 | 173 | 7 | 179 | ChainA, ArnB aminotransferase [Salmonella enterica subsp. enterica serovar Typhimurium],1MDZ_A Chain A, ArnB aminotransferase [Salmonella enterica subsp. enterica serovar Typhimurium] |
| 1MDO_A | 5.86e-88 | 2 | 173 | 8 | 179 | ChainA, ArnB aminotransferase [Salmonella enterica subsp. enterica serovar Typhimurium] |
| 4LC3_A | 4.65e-39 | 4 | 172 | 16 | 184 | X-raycrystal structure of a putative UDP-4-amino-4-deoxy-l-arabinose--oxoglutarate aminotransferase from Burkholderia cenocepacia [Burkholderia cenocepacia J2315],4LC3_B X-ray crystal structure of a putative UDP-4-amino-4-deoxy-l-arabinose--oxoglutarate aminotransferase from Burkholderia cenocepacia [Burkholderia cenocepacia J2315] |
| 3NU7_A | 6.32e-37 | 18 | 166 | 20 | 168 | WbpE,an Aminotransferase from Pseudomonas aeruginosa Involved in O-antigen Assembly in Complex with the Cofactor PMP [Pseudomonas aeruginosa],3NU7_B WbpE, an Aminotransferase from Pseudomonas aeruginosa Involved in O-antigen Assembly in Complex with the Cofactor PMP [Pseudomonas aeruginosa],3NUB_A WbpE, an Aminotransferase from Pseudomonas aeruginosa Involved in O-antigen Assembly in Complex with Product as the External Aldimine [Pseudomonas aeruginosa],3NUB_B WbpE, an Aminotransferase from Pseudomonas aeruginosa Involved in O-antigen Assembly in Complex with Product as the External Aldimine [Pseudomonas aeruginosa] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B7LM77 | 3.34e-162 | 139 | 446 | 33 | 325 | Undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase OS=Escherichia fergusonii (strain ATCC 35469 / DSM 13698 / CCUG 18766 / IAM 14443 / JCM 21226 / LMG 7866 / NBRC 102419 / NCTC 12128 / CDC 0568-73) OX=585054 GN=arnC PE=3 SV=1 |
| A4WAM4 | 4.00e-161 | 139 | 447 | 33 | 326 | Undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase OS=Enterobacter sp. (strain 638) OX=399742 GN=arnC PE=3 SV=1 |
| A6TF99 | 5.67e-161 | 139 | 445 | 33 | 325 | Undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase OS=Klebsiella pneumoniae subsp. pneumoniae (strain ATCC 700721 / MGH 78578) OX=272620 GN=arnC PE=3 SV=1 |
| B5XTK8 | 2.30e-160 | 139 | 445 | 33 | 325 | Undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase OS=Klebsiella pneumoniae (strain 342) OX=507522 GN=arnC PE=3 SV=1 |
| B7M5T6 | 2.22e-159 | 139 | 447 | 33 | 322 | Undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase OS=Escherichia coli O8 (strain IAI1) OX=585034 GN=arnC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
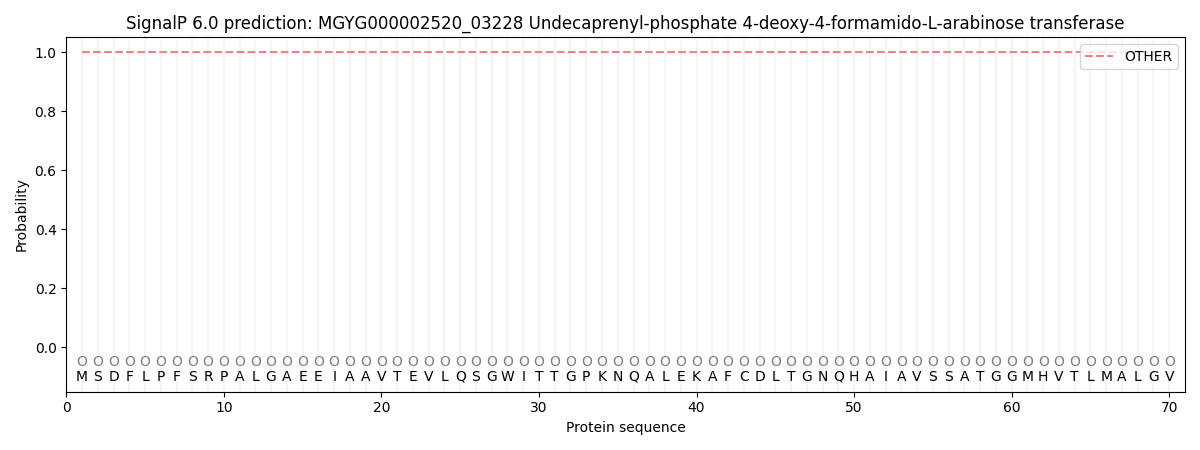
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000031 | 0.000002 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |