You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002520_03503
You are here: Home > Sequence: MGYG000002520_03503
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pluralibacter gergoviae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Pluralibacter; Pluralibacter gergoviae | |||||||||||
| CAZyme ID | MGYG000002520_03503 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3788339; End: 3791113 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 1.10e-47 | 739 | 914 | 18 | 189 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| smart00047 | LYZ2 | 2.60e-12 | 815 | 915 | 3 | 102 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| pfam01832 | Glucosaminidase | 8.91e-07 | 826 | 914 | 2 | 80 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVR04460.1 | 0.0 | 115 | 924 | 1 | 810 |
| QXC97730.1 | 0.0 | 1 | 913 | 1 | 910 |
| BBW76016.1 | 0.0 | 1 | 913 | 1 | 910 |
| AYL66980.1 | 0.0 | 1 | 915 | 1 | 918 |
| QFU62461.1 | 0.0 | 1 | 913 | 1 | 918 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4PI7_A | 9.38e-28 | 764 | 914 | 16 | 173 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI8_A | 5.92e-27 | 764 | 914 | 16 | 173 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 6FXP_A | 6.94e-26 | 739 | 917 | 14 | 193 | ChainA, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50],6FXP_B Chain B, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50] |
| 6U0O_B | 7.10e-26 | 739 | 917 | 44 | 223 | ChainB, LYZ2 domain-containing protein [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 6FXO_A | 5.38e-24 | 739 | 915 | 6 | 190 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P39848 | 1.38e-24 | 770 | 913 | 678 | 823 | Beta-N-acetylglucosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=lytD PE=1 SV=1 |
| O33635 | 2.04e-22 | 521 | 915 | 924 | 1281 | Bifunctional autolysin OS=Staphylococcus epidermidis OX=1282 GN=atl PE=1 SV=1 |
| Q8CPQ1 | 2.04e-22 | 521 | 915 | 924 | 1281 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=atl PE=3 SV=1 |
| Q5HQB9 | 2.04e-22 | 521 | 915 | 924 | 1281 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=atl PE=3 SV=1 |
| Q6GI31 | 4.00e-21 | 739 | 915 | 1019 | 1203 | Bifunctional autolysin OS=Staphylococcus aureus (strain MRSA252) OX=282458 GN=atl PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
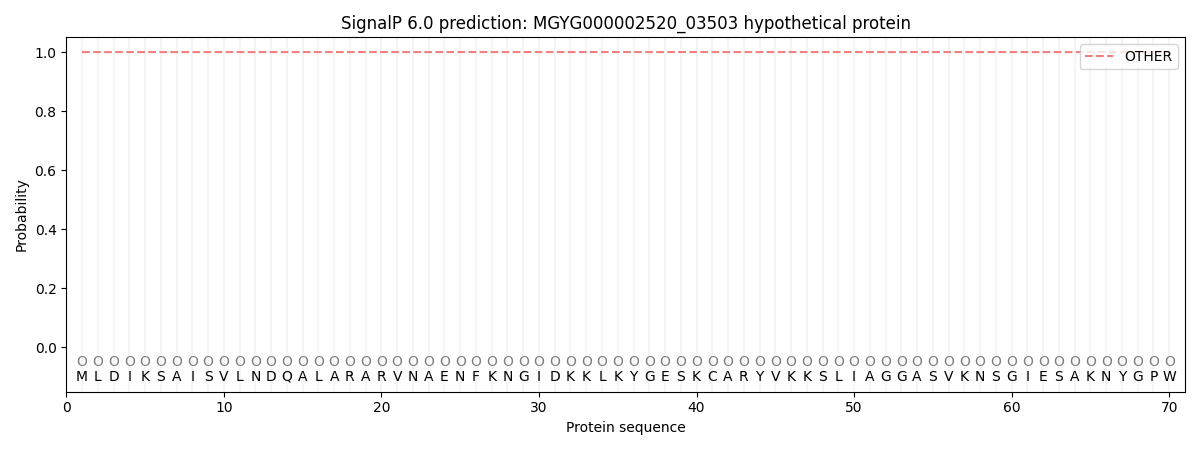
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000072 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
