You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002526_02966
You are here: Home > Sequence: MGYG000002526_02966
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aeromonas hydrophila | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Aeromonadaceae; Aeromonas; Aeromonas hydrophila | |||||||||||
| CAZyme ID | MGYG000002526_02966 | |||||||||||
| CAZy Family | GH20 | |||||||||||
| CAZyme Description | N,N'-diacetylchitobiase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3267000; End: 3269663 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH20 | 328 | 768 | 3.1e-99 | 0.973293768545994 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06569 | GH20_Sm-chitobiase-like | 0.0 | 329 | 804 | 1 | 445 | The chitobiase of Serratia marcescens is a beta-N-1,4-acetylhexosaminidase with a glycosyl hydrolase family 20 (GH20) domain that hydrolyzes the beta-1,4-glycosidic linkages in oligomers derived from chitin. Chitin is degraded by a two step process: i) a chitinase hydrolyzes the chitin to oligosaccharides and disaccharides such as di-N-acetyl-D-glucosamine and chitobiose, ii) chitobiase then further degrades these oligomers into monomers. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
| COG3525 | Chb | 2.07e-141 | 57 | 884 | 8 | 732 | N-acetyl-beta-hexosaminidase [Carbohydrate transport and metabolism]. |
| pfam00728 | Glyco_hydro_20 | 2.53e-135 | 333 | 768 | 1 | 345 | Glycosyl hydrolase family 20, catalytic domain. This domain has a TIM barrel fold. |
| cd06563 | GH20_chitobiase-like | 2.40e-104 | 333 | 768 | 1 | 344 | The chitobiase of Serratia marcescens is a beta-N-1,4-acetylhexosaminidase with a glycosyl hydrolase family 20 (GH20) domain that hydrolyzes the beta-1,4-glycosidic linkages in oligomers derived from chitin. Chitin is degraded by a two step process: i) a chitinase hydrolyzes the chitin to oligosaccharides and disaccharides such as di-N-acetyl-D-glucosamine and chitobiose, ii) chitobiase then further degrades these oligomers into monomers. This GH20 domain family includes an N-acetylglucosamidase (GlcNAcase A) from Pseudoalteromonas piscicida and an N-acetylhexosaminidase (SpHex) from Streptomyces plicatus. SpHex lacks the C-terminal PKD (polycystic kidney disease I)-like domain found in the chitobiases. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
| pfam03173 | CHB_HEX | 7.82e-79 | 38 | 196 | 1 | 152 | Putative carbohydrate binding domain. This domain represents the N terminal domain in chitobiases and beta-hexosaminidases EC:3.2.1.52. It is composed of a beta sandwich structure that is similar in structure to the cellulose binding domain of cellulase from Cellulomonas fimi. This suggests that this may be a carbohydrate binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANT68580.1 | 0.0 | 1 | 887 | 1 | 887 |
| AWA06002.1 | 0.0 | 1 | 887 | 1 | 887 |
| QWL69183.1 | 0.0 | 1 | 887 | 1 | 887 |
| QBX72174.1 | 0.0 | 1 | 887 | 1 | 887 |
| AVP83932.1 | 0.0 | 1 | 887 | 1 | 887 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1QBA_A | 0.0 | 28 | 886 | 2 | 858 | BACTERIALCHITOBIASE, GLYCOSYL HYDROLASE FAMILY 20 [Serratia marcescens],1QBB_A BACTERIAL CHITOBIASE COMPLEXED WITH CHITOBIOSE (DINAG) [Serratia marcescens] |
| 1C7T_A | 8.62e-319 | 28 | 886 | 2 | 858 | ChainA, BETA-N-ACETYLHEXOSAMINIDASE [Serratia marcescens] |
| 1C7S_A | 4.31e-318 | 28 | 886 | 2 | 858 | ChainA, BETA-N-ACETYLHEXOSAMINIDASE [Serratia marcescens] |
| 6EZR_A | 3.93e-51 | 329 | 820 | 259 | 638 | Crystalstructure of GH20 Exo beta-N-Acetylglucosaminidase from Vibrio harveyi [Vibrio harveyi],6EZR_B Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase from Vibrio harveyi [Vibrio harveyi],6EZS_A Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase from Vibrio harveyi in complex with N-acetylglucosamine [Vibrio harveyi],6EZS_B Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase from Vibrio harveyi in complex with N-acetylglucosamine [Vibrio harveyi],6K35_A Crystal structure of GH20 exo beta-N-acetylglucosaminidase from Vibrio harveyi in complex with NAG-thiazoline [Vibrio harveyi],6K35_B Crystal structure of GH20 exo beta-N-acetylglucosaminidase from Vibrio harveyi in complex with NAG-thiazoline [Vibrio harveyi] |
| 6EZT_A | 4.26e-50 | 329 | 820 | 256 | 635 | Crystalstructure of GH20 Exo beta-N-Acetylglucosaminidase D437A inactive mutant from Vibrio harveyi [Vibrio harveyi],6EZT_B Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase D437A inactive mutant from Vibrio harveyi [Vibrio harveyi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q54468 | 0.0 | 26 | 886 | 27 | 885 | Chitobiase OS=Serratia marcescens OX=615 GN=chb PE=1 SV=1 |
| P13670 | 0.0 | 3 | 886 | 2 | 882 | N,N'-diacetylchitobiase OS=Vibrio harveyi OX=669 GN=chb PE=1 SV=1 |
| Q04786 | 5.80e-207 | 31 | 886 | 10 | 844 | Beta-hexosaminidase OS=Vibrio vulnificus OX=672 GN=hex PE=3 SV=1 |
| P49007 | 1.18e-133 | 28 | 805 | 22 | 768 | Beta-hexosaminidase B OS=Pseudoalteromonas piscicida OX=43662 GN=nag096 PE=3 SV=1 |
| H2A0L6 | 1.14e-45 | 100 | 854 | 104 | 870 | Putative beta-hexosaminidase OS=Margaritifera margaritifera OX=102329 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
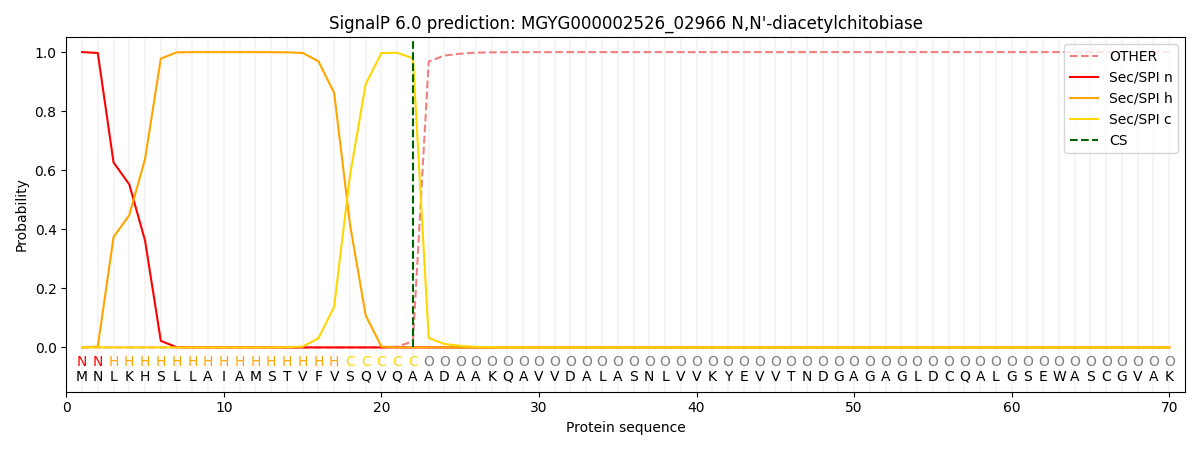
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000220 | 0.999138 | 0.000154 | 0.000163 | 0.000149 | 0.000136 |
