You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002529_03322
You are here: Home > Sequence: MGYG000002529_03322
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aeromonas veronii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Aeromonadaceae; Aeromonas; Aeromonas veronii | |||||||||||
| CAZyme ID | MGYG000002529_03322 | |||||||||||
| CAZy Family | CBM5 | |||||||||||
| CAZyme Description | Metalloprotease StcE | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 46862; End: 49903 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam10462 | Peptidase_M66 | 1.46e-89 | 255 | 556 | 1 | 306 | Peptidase M66. This family of metallopeptidases contains StcE, a virulence factor found in Shiga toxigenic Escherichia coli organisms. StcE peptidase cleaves C1 esterase inhibitor. |
| pfam12561 | TagA | 2.34e-46 | 685 | 780 | 1 | 99 | ToxR activated gene A lipoprotein. This domain family is found in bacteria, and is approximately 140 amino acids in length. The family is found in association with pfam10462. There is a conserved GAG sequence motif. This family is a bacterial lipoprotein. |
| cd12215 | ChiC_BD | 2.26e-16 | 919 | 960 | 1 | 42 | Chitin-binding domain of chitinase C. Chitin-binding domain of chitinase C (ChiC) of Streptomyces griseus and related proteins. Chitinase C is a family 19 chitinase, and consists of a N-terminal chitin binding domain and a C-terminal chitin-catalytic domain that effects degradation. Chitinases function in invertebrates in the degradation of old exoskeletons, in fungi to utilize chitin in cell walls, and in bacteria which use chitin as an energy source. ChiC contains the characteristic chitin-binding aromatic residues. |
| smart00495 | ChtBD3 | 6.82e-15 | 918 | 958 | 1 | 41 | Chitin-binding domain type 3. |
| cd12215 | ChiC_BD | 8.67e-14 | 968 | 1004 | 3 | 40 | Chitin-binding domain of chitinase C. Chitin-binding domain of chitinase C (ChiC) of Streptomyces griseus and related proteins. Chitinase C is a family 19 chitinase, and consists of a N-terminal chitin binding domain and a C-terminal chitin-catalytic domain that effects degradation. Chitinases function in invertebrates in the degradation of old exoskeletons, in fungi to utilize chitin in cell walls, and in bacteria which use chitin as an energy source. ChiC contains the characteristic chitin-binding aromatic residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AYV36206.1 | 0.0 | 1 | 1013 | 1 | 1013 |
| QET81665.1 | 0.0 | 1 | 1013 | 1 | 1013 |
| QHC10148.1 | 0.0 | 1 | 1013 | 1 | 1013 |
| QMS78739.1 | 0.0 | 1 | 1013 | 1 | 1013 |
| BBT94192.1 | 0.0 | 1 | 1013 | 1 | 1013 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3UJZ_A | 0.0 | 50 | 820 | 15 | 783 | Crystalstructure of enterohemorrhagic E. coli StcE [Escherichia coli O157:H7] |
| 4DNY_A | 6.22e-46 | 138 | 257 | 7 | 126 | Crystalstructure of enterohemorrhagic E. coli StcE(132-251) [Escherichia coli O157:H7] |
| 4TXG_A | 1.46e-07 | 910 | 963 | 45 | 98 | CrystalStructure of a Family GH18 Chitinase from Chromobacterium violaceum [Chromobacterium violaceum ATCC 12472] |
| 4HMC_A | 9.53e-06 | 813 | 956 | 392 | 523 | Crystalstructure of cold-adapted chitinase from Moritella marina [Moritella marina],4HMD_A Crystal structure of cold-adapted chitinase from Moritella marina with a reaction intermediate - oxazolinium ion (NGO) [Moritella marina],4HME_A Crystal structure of cold-adapted chitinase from Moritella marina with a reaction product - NAG2 [Moritella marina] |
| 4MB3_A | 9.53e-06 | 813 | 956 | 392 | 523 | Crystalstructure of E153Q mutant of cold-adapted chitinase from Moritella marina [Moritella marina],4MB4_A Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag4 [Moritella marina],4MB5_A Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag5 [Moritella marina] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O82882 | 0.0 | 13 | 820 | 3 | 812 | Metalloprotease StcE OS=Escherichia coli O157:H7 OX=83334 GN=stcE PE=1 SV=2 |
| P0C6Q7 | 2.57e-58 | 255 | 556 | 231 | 535 | ToxR-activated gene A lipoprotein OS=Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961) OX=243277 GN=tagA PE=3 SV=1 |
| A5F398 | 8.27e-58 | 255 | 556 | 231 | 535 | ToxR-activated gene A lipoprotein OS=Vibrio cholerae serotype O1 (strain ATCC 39541 / Classical Ogawa 395 / O395) OX=345073 GN=tagA PE=3 SV=2 |
| P32823 | 7.38e-18 | 813 | 962 | 685 | 819 | Chitinase A OS=Pseudoalteromonas piscicida OX=43662 GN=chiA PE=1 SV=1 |
| Q3JLY2 | 5.51e-17 | 255 | 583 | 192 | 527 | Dictomallein OS=Burkholderia pseudomallei (strain 1710b) OX=320372 GN=dtmL PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
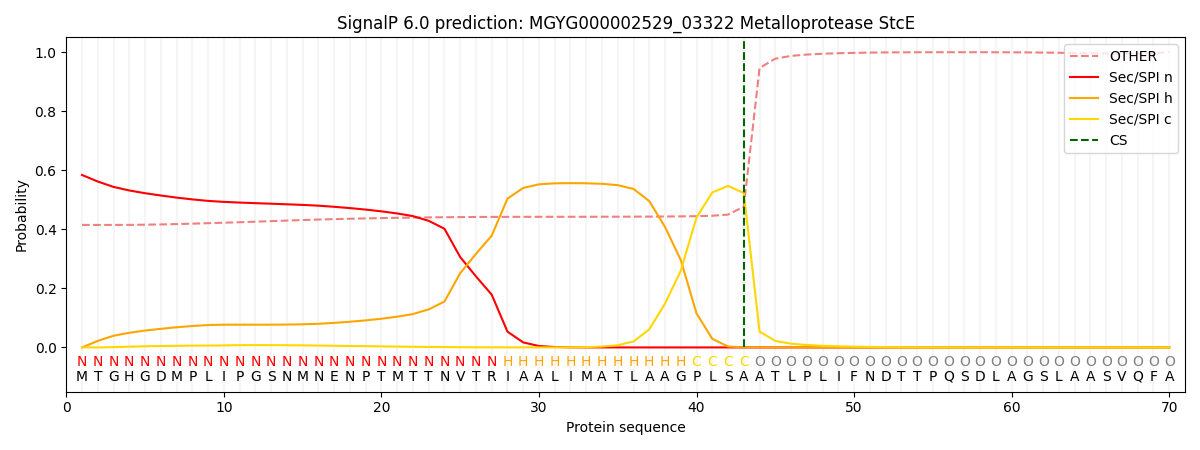
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.431330 | 0.564405 | 0.002186 | 0.000829 | 0.000501 | 0.000735 |

