You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002533_01806
You are here: Home > Sequence: MGYG000002533_01806
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Vibrio vulnificus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Vibrionaceae; Vibrio; Vibrio vulnificus | |||||||||||
| CAZyme ID | MGYG000002533_01806 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | Cytolysin | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 484523; End: 485938 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07968 | Leukocidin | 1.45e-74 | 72 | 331 | 1 | 250 | Leukocidin/Hemolysin toxin family. |
| smart00458 | RICIN | 2.09e-17 | 343 | 465 | 1 | 118 | Ricin-type beta-trefoil. Carbohydrate-binding domain formed from presumed gene triplication. |
| cd00161 | RICIN | 2.35e-15 | 340 | 463 | 1 | 123 | Ricin-type beta-trefoil; Carbohydrate-binding domain formed from presumed gene triplication. The domain is found in a variety of molecules serving diverse functions such as enzymatic activity, inhibitory toxicity and signal transduction. Highly specific ligand binding occurs on exposed surfaces of the compact domain sturcture. |
| pfam00652 | Ricin_B_lectin | 2.86e-08 | 338 | 430 | 1 | 94 | Ricin-type beta-trefoil lectin domain. |
| pfam00652 | Ricin_B_lectin | 4.82e-07 | 317 | 420 | 32 | 126 | Ricin-type beta-trefoil lectin domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAC99982.1 | 0.0 | 1 | 471 | 1 | 471 |
| AMG10720.1 | 0.0 | 1 | 471 | 1 | 471 |
| AUJ37047.1 | 0.0 | 1 | 471 | 1 | 471 |
| ADV88903.1 | 0.0 | 1 | 471 | 1 | 471 |
| ARN68290.1 | 0.0 | 1 | 471 | 1 | 471 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4OWJ_A | 4.90e-95 | 336 | 471 | 3 | 138 | CrystalStructure of the Vibrio vulnificus Hemolysin/Cytolysin Beta-Trefoil Lectin [Vibrio vulnificus CMCP6],4OWJ_B Crystal Structure of the Vibrio vulnificus Hemolysin/Cytolysin Beta-Trefoil Lectin [Vibrio vulnificus CMCP6],4OWJ_C Crystal Structure of the Vibrio vulnificus Hemolysin/Cytolysin Beta-Trefoil Lectin [Vibrio vulnificus CMCP6],4OWJ_D Crystal Structure of the Vibrio vulnificus Hemolysin/Cytolysin Beta-Trefoil Lectin [Vibrio vulnificus CMCP6],4OWJ_E Crystal Structure of the Vibrio vulnificus Hemolysin/Cytolysin Beta-Trefoil Lectin [Vibrio vulnificus CMCP6],4OWJ_F Crystal Structure of the Vibrio vulnificus Hemolysin/Cytolysin Beta-Trefoil Lectin [Vibrio vulnificus CMCP6],4OWJ_G Crystal Structure of the Vibrio vulnificus Hemolysin/Cytolysin Beta-Trefoil Lectin [Vibrio vulnificus CMCP6],4OWK_A Crystal Structure of the Vibrio vulnificus Hemolysin/Cytolysin Beta-Trefoil Lectin with N-Acetyl-D-Galactosamine Bound [Vibrio vulnificus CMCP6],4OWK_B Crystal Structure of the Vibrio vulnificus Hemolysin/Cytolysin Beta-Trefoil Lectin with N-Acetyl-D-Galactosamine Bound [Vibrio vulnificus CMCP6],4OWK_C Crystal Structure of the Vibrio vulnificus Hemolysin/Cytolysin Beta-Trefoil Lectin with N-Acetyl-D-Galactosamine Bound [Vibrio vulnificus CMCP6],4OWK_D Crystal Structure of the Vibrio vulnificus Hemolysin/Cytolysin Beta-Trefoil Lectin with N-Acetyl-D-Galactosamine Bound [Vibrio vulnificus CMCP6],4OWK_E Crystal Structure of the Vibrio vulnificus Hemolysin/Cytolysin Beta-Trefoil Lectin with N-Acetyl-D-Galactosamine Bound [Vibrio vulnificus CMCP6],4OWK_F Crystal Structure of the Vibrio vulnificus Hemolysin/Cytolysin Beta-Trefoil Lectin with N-Acetyl-D-Galactosamine Bound [Vibrio vulnificus CMCP6],4OWK_G Crystal Structure of the Vibrio vulnificus Hemolysin/Cytolysin Beta-Trefoil Lectin with N-Acetyl-D-Galactosamine Bound [Vibrio vulnificus CMCP6],4OWL_A Crystal Structure of the Vibrio vulnificus Hemolysin/Cytolysin Beta-Trefoil Lectin with N-Acetyl-D-Lactosamine Bound [Vibrio vulnificus CMCP6],4OWL_B Crystal Structure of the Vibrio vulnificus Hemolysin/Cytolysin Beta-Trefoil Lectin with N-Acetyl-D-Lactosamine Bound [Vibrio vulnificus CMCP6],4OWL_C Crystal Structure of the Vibrio vulnificus Hemolysin/Cytolysin Beta-Trefoil Lectin with N-Acetyl-D-Lactosamine Bound [Vibrio vulnificus CMCP6],4OWL_D Crystal Structure of the Vibrio vulnificus Hemolysin/Cytolysin Beta-Trefoil Lectin with N-Acetyl-D-Lactosamine Bound [Vibrio vulnificus CMCP6],4OWL_E Crystal Structure of the Vibrio vulnificus Hemolysin/Cytolysin Beta-Trefoil Lectin with N-Acetyl-D-Lactosamine Bound [Vibrio vulnificus CMCP6],4OWL_F Crystal Structure of the Vibrio vulnificus Hemolysin/Cytolysin Beta-Trefoil Lectin with N-Acetyl-D-Lactosamine Bound [Vibrio vulnificus CMCP6],4OWL_G Crystal Structure of the Vibrio vulnificus Hemolysin/Cytolysin Beta-Trefoil Lectin with N-Acetyl-D-Lactosamine Bound [Vibrio vulnificus CMCP6] |
| 3O44_A | 4.72e-33 | 51 | 422 | 48 | 411 | CrystalStructure of the Vibrio cholerae Cytolysin (HlyA) Heptameric Pore [Vibrio cholerae 12129(1)],3O44_B Crystal Structure of the Vibrio cholerae Cytolysin (HlyA) Heptameric Pore [Vibrio cholerae 12129(1)],3O44_C Crystal Structure of the Vibrio cholerae Cytolysin (HlyA) Heptameric Pore [Vibrio cholerae 12129(1)],3O44_D Crystal Structure of the Vibrio cholerae Cytolysin (HlyA) Heptameric Pore [Vibrio cholerae 12129(1)],3O44_E Crystal Structure of the Vibrio cholerae Cytolysin (HlyA) Heptameric Pore [Vibrio cholerae 12129(1)],3O44_F Crystal Structure of the Vibrio cholerae Cytolysin (HlyA) Heptameric Pore [Vibrio cholerae 12129(1)],3O44_G Crystal Structure of the Vibrio cholerae Cytolysin (HlyA) Heptameric Pore [Vibrio cholerae 12129(1)],3O44_H Crystal Structure of the Vibrio cholerae Cytolysin (HlyA) Heptameric Pore [Vibrio cholerae 12129(1)],3O44_I Crystal Structure of the Vibrio cholerae Cytolysin (HlyA) Heptameric Pore [Vibrio cholerae 12129(1)],3O44_J Crystal Structure of the Vibrio cholerae Cytolysin (HlyA) Heptameric Pore [Vibrio cholerae 12129(1)],3O44_K Crystal Structure of the Vibrio cholerae Cytolysin (HlyA) Heptameric Pore [Vibrio cholerae 12129(1)],3O44_L Crystal Structure of the Vibrio cholerae Cytolysin (HlyA) Heptameric Pore [Vibrio cholerae 12129(1)],3O44_M Crystal Structure of the Vibrio cholerae Cytolysin (HlyA) Heptameric Pore [Vibrio cholerae 12129(1)],3O44_N Crystal Structure of the Vibrio cholerae Cytolysin (HlyA) Heptameric Pore [Vibrio cholerae 12129(1)] |
| 1XEZ_A | 8.57e-33 | 51 | 422 | 176 | 539 | CrystalStructure Of The Vibrio Cholerae Cytolysin (HlyA) Pro-Toxin With Octylglucoside Bound [Vibrio cholerae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P19247 | 0.0 | 1 | 471 | 1 | 471 | Cytolysin OS=Vibrio vulnificus (strain CMCP6) OX=216895 GN=vvhA PE=1 SV=2 |
| P55870 | 6.17e-39 | 60 | 425 | 213 | 569 | Hemolysin ahh1 OS=Aeromonas hydrophila subsp. hydrophila (strain ATCC 7966 / DSM 30187 / BCRC 13018 / CCUG 14551 / JCM 1027 / KCTC 2358 / NCIMB 9240 / NCTC 8049) OX=380703 GN=ahh1 PE=3 SV=2 |
| P09545 | 1.50e-32 | 51 | 422 | 196 | 559 | Hemolysin OS=Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961) OX=243277 GN=hlyA PE=1 SV=2 |
| Q08677 | 4.48e-18 | 60 | 425 | 171 | 526 | Hemolysin 4 OS=Aeromonas salmonicida OX=645 GN=ash4 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
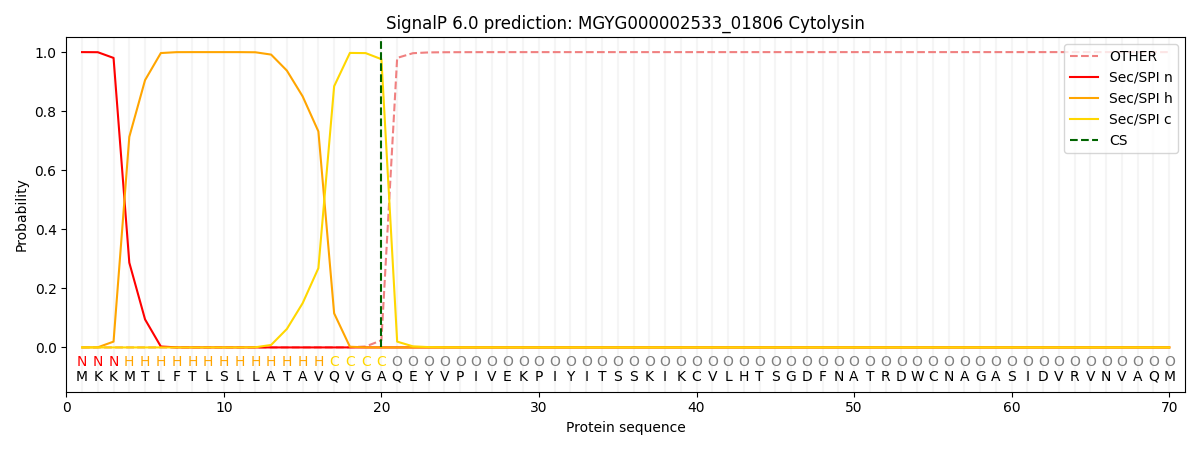
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000249 | 0.999087 | 0.000163 | 0.000165 | 0.000155 | 0.000144 |
