You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002533_03268
You are here: Home > Sequence: MGYG000002533_03268
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Vibrio vulnificus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Vibrionaceae; Vibrio; Vibrio vulnificus | |||||||||||
| CAZyme ID | MGYG000002533_03268 | |||||||||||
| CAZy Family | PL9 | |||||||||||
| CAZyme Description | Pectate disaccharide-lyase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 135161; End: 137392 Strand: - | |||||||||||
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAC97406.1 | 0.0 | 1 | 743 | 1 | 743 |
| ANH66082.1 | 0.0 | 1 | 743 | 1 | 743 |
| ALM73720.1 | 0.0 | 1 | 743 | 1 | 743 |
| ARN68785.1 | 0.0 | 1 | 743 | 1 | 743 |
| AMG10357.1 | 0.0 | 1 | 743 | 1 | 743 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1RU4_A | 9.69e-32 | 384 | 709 | 18 | 347 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P22751 | 1.16e-204 | 19 | 695 | 25 | 688 | Pectate disaccharide-lyase OS=Dickeya chrysanthemi OX=556 GN=pelX PE=1 SV=1 |
| P0C1A7 | 7.96e-31 | 384 | 709 | 43 | 372 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
| P0C1A6 | 4.80e-30 | 384 | 709 | 43 | 372 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
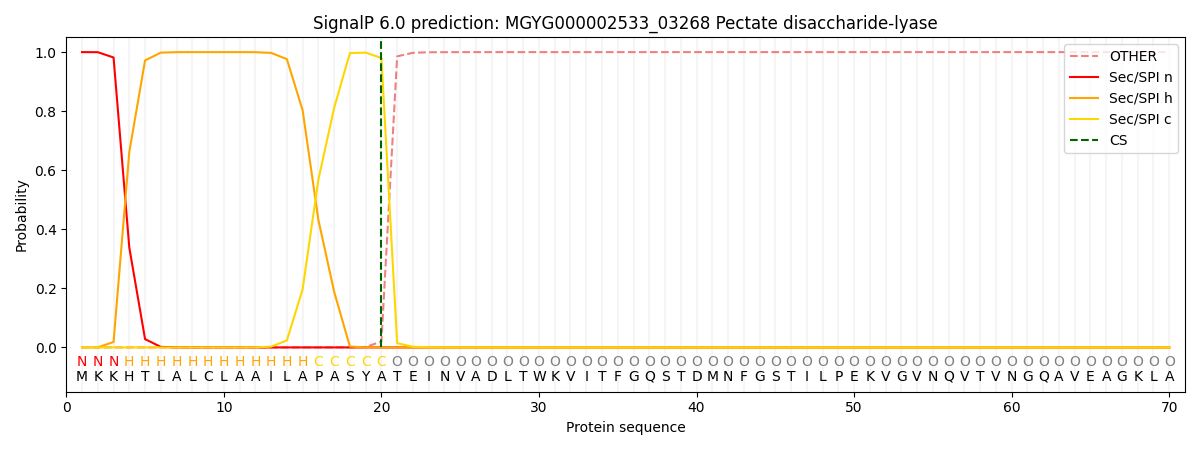
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000237 | 0.999025 | 0.000190 | 0.000181 | 0.000174 | 0.000158 |
