You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002533_03272
You are here: Home > Sequence: MGYG000002533_03272
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Vibrio vulnificus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Vibrionaceae; Vibrio; Vibrio vulnificus | |||||||||||
| CAZyme ID | MGYG000002533_03272 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 141870; End: 142337 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 27 | 132 | 1.2e-18 | 0.8225806451612904 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 3.09e-10 | 26 | 136 | 5 | 122 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam17132 | Glyco_hydro_106 | 4.62e-04 | 52 | 124 | 206 | 282 | alpha-L-rhamnosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADV89318.1 | 1.40e-110 | 1 | 155 | 1 | 155 |
| ANH66086.1 | 1.40e-110 | 1 | 155 | 1 | 155 |
| ARN68789.1 | 1.40e-110 | 1 | 155 | 1 | 155 |
| ALM73724.1 | 1.40e-110 | 1 | 155 | 1 | 155 |
| AIL73346.1 | 1.40e-110 | 1 | 155 | 1 | 155 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2JD9_A | 6.59e-25 | 24 | 145 | 12 | 135 | Structureof a pectin binding carbohydrate binding module determined in an orthorhombic crystal form. [Yersinia enterocolitica],2JDA_A Structure of a pectin binding carbohydrate binding module determined in an monoclinic crystal form. [Yersinia enterocolitica],2JDA_B Structure of a pectin binding carbohydrate binding module determined in an monoclinic crystal form. [Yersinia enterocolitica] |
| 7D29_A | 5.03e-15 | 36 | 141 | 24 | 129 | CBM32of AlyQ [Persicobacter sp. CCB-QB2],7D2A_A CBM32 of AlyQ in complex with 4,5-unsaturated mannuronic acid [Persicobacter sp. CCB-QB2] |
| 5XNR_A | 6.63e-14 | 36 | 141 | 24 | 129 | TruncatedAlyQ with CBM32 and alginate lyase domains [Persicobacter sp. CCB-QB2] |
| 5ZU6_A | 6.30e-13 | 36 | 136 | 47 | 149 | ACBM32 derived from alginate lyase B (AlyB-OU02) [Vibrio] |
| 5ZU5_A | 4.07e-12 | 36 | 136 | 47 | 149 | Crystalstructure of a full length alginate lyase with CBM domain [Vibrio splendidus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
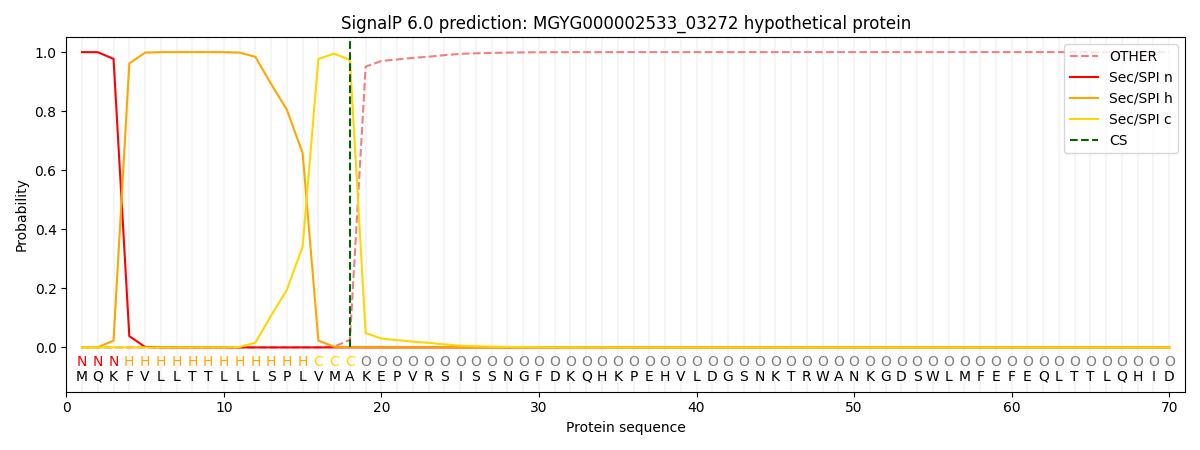
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000350 | 0.998887 | 0.000206 | 0.000175 | 0.000173 | 0.000154 |
