You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002534_00200
You are here: Home > Sequence: MGYG000002534_00200
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
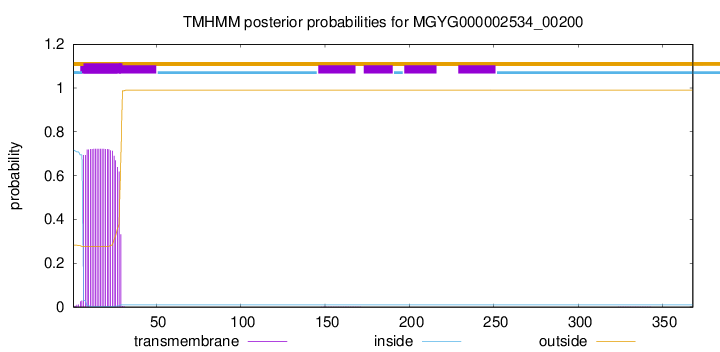
TMHMM annotations
Basic Information help
| Species | Citrobacter_A farmeri | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Citrobacter_A; Citrobacter_A farmeri | |||||||||||
| CAZyme ID | MGYG000002534_00200 | |||||||||||
| CAZy Family | GH8 | |||||||||||
| CAZyme Description | Endoglucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 213808; End: 214914 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK11097 | PRK11097 | 0.0 | 1 | 367 | 1 | 375 | cellulase. |
| COG3405 | BcsZ | 3.94e-179 | 1 | 366 | 2 | 360 | Endo-1,4-beta-D-glucanase Y [Carbohydrate transport and metabolism]. |
| pfam01270 | Glyco_hydro_8 | 3.90e-132 | 22 | 346 | 1 | 321 | Glycosyl hydrolases family 8. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AST77768.1 | 8.01e-280 | 1 | 368 | 2 | 369 |
| QZE45012.1 | 3.01e-277 | 1 | 368 | 1 | 368 |
| AKE60524.1 | 3.37e-274 | 1 | 368 | 1 | 368 |
| AMG92715.1 | 1.37e-273 | 1 | 368 | 1 | 368 |
| AUO66932.1 | 1.43e-273 | 1 | 368 | 2 | 369 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3QXQ_A | 1.45e-232 | 22 | 368 | 1 | 347 | Structureof the bacterial cellulose synthase subunit Z in complex with cellopentaose [Escherichia coli K-12],3QXQ_B Structure of the bacterial cellulose synthase subunit Z in complex with cellopentaose [Escherichia coli K-12],3QXQ_C Structure of the bacterial cellulose synthase subunit Z in complex with cellopentaose [Escherichia coli K-12],3QXQ_D Structure of the bacterial cellulose synthase subunit Z in complex with cellopentaose [Escherichia coli K-12] |
| 3QXF_A | 8.94e-227 | 22 | 368 | 1 | 347 | Structureof the bacterial cellulose synthase subunit Z [Escherichia coli K-12],3QXF_B Structure of the bacterial cellulose synthase subunit Z [Escherichia coli K-12],3QXF_C Structure of the bacterial cellulose synthase subunit Z [Escherichia coli K-12],3QXF_D Structure of the bacterial cellulose synthase subunit Z [Escherichia coli K-12] |
| 7F81_A | 2.52e-209 | 22 | 367 | 5 | 350 | ChainA, Glucanase [Enterobacter sp. CJF-002],7F81_B Chain B, Glucanase [Enterobacter sp. CJF-002],7F81_C Chain C, Glucanase [Enterobacter sp. CJF-002],7F81_D Chain D, Glucanase [Enterobacter sp. CJF-002] |
| 7F82_A | 4.17e-208 | 22 | 367 | 5 | 350 | ChainA, Glucanase [Enterobacter sp. CJF-002],7F82_B Chain B, Glucanase [Enterobacter sp. CJF-002],7F82_C Chain C, Glucanase [Enterobacter sp. CJF-002],7F82_D Chain D, Glucanase [Enterobacter sp. CJF-002] |
| 4Q2B_A | 2.44e-128 | 25 | 365 | 4 | 348 | Thecrystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440 [Pseudomonas putida KT2440],4Q2B_B The crystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440 [Pseudomonas putida KT2440],4Q2B_C The crystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440 [Pseudomonas putida KT2440],4Q2B_D The crystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440 [Pseudomonas putida KT2440],4Q2B_E The crystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440 [Pseudomonas putida KT2440],4Q2B_F The crystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440 [Pseudomonas putida KT2440] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8ZLB7 | 8.68e-244 | 1 | 367 | 2 | 368 | Endoglucanase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=bcsZ PE=3 SV=1 |
| Q8Z289 | 2.49e-243 | 1 | 367 | 2 | 368 | Endoglucanase OS=Salmonella typhi OX=90370 GN=bcsZ PE=3 SV=1 |
| Q8X5L9 | 3.40e-243 | 1 | 368 | 1 | 368 | Endoglucanase OS=Escherichia coli O157:H7 OX=83334 GN=bcsZ PE=3 SV=1 |
| P37651 | 2.79e-242 | 1 | 368 | 1 | 368 | Endoglucanase OS=Escherichia coli (strain K12) OX=83333 GN=bcsZ PE=1 SV=1 |
| P58935 | 3.51e-108 | 6 | 360 | 13 | 375 | Endoglucanase OS=Xanthomonas axonopodis pv. citri (strain 306) OX=190486 GN=bcsZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
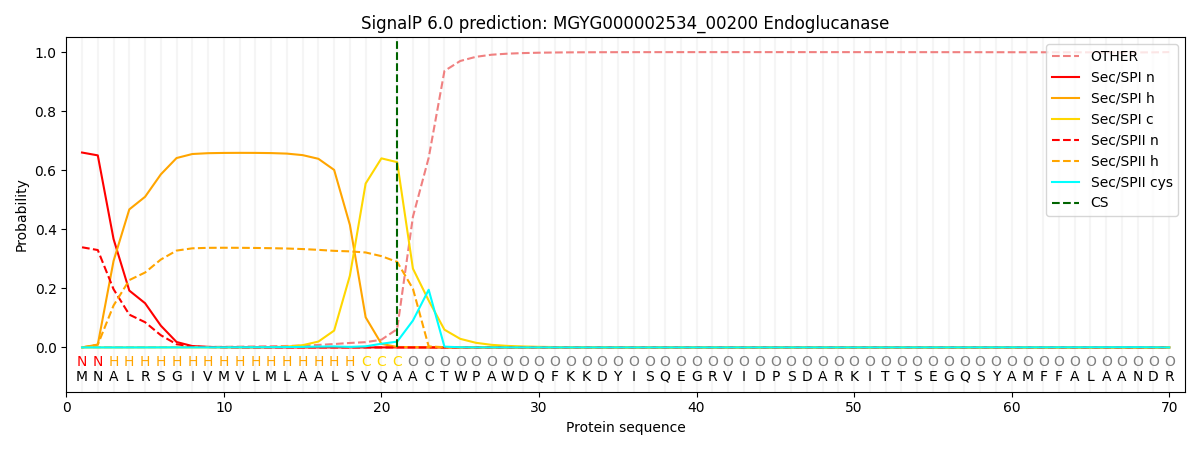
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002261 | 0.652271 | 0.344689 | 0.000258 | 0.000237 | 0.000256 |