You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002540_00177
You are here: Home > Sequence: MGYG000002540_00177
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paraprevotella clara | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Paraprevotella; Paraprevotella clara | |||||||||||
| CAZyme ID | MGYG000002540_00177 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 15871; End: 18678 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM13 | 402 | 538 | 3.9e-18 | 0.7074468085106383 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13385 | Laminin_G_3 | 3.41e-29 | 682 | 836 | 1 | 151 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| pfam14200 | RicinB_lectin_2 | 1.45e-13 | 437 | 521 | 4 | 88 | Ricin-type beta-trefoil lectin domain-like. |
| pfam00652 | Ricin_B_lectin | 2.41e-08 | 455 | 539 | 4 | 88 | Ricin-type beta-trefoil lectin domain. |
| pfam14200 | RicinB_lectin_2 | 1.00e-07 | 398 | 481 | 7 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 1.24e-07 | 487 | 539 | 1 | 58 | Ricin-type beta-trefoil lectin domain-like. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ60208.1 | 0.0 | 2 | 934 | 13 | 940 |
| QUT88791.1 | 0.0 | 3 | 934 | 14 | 940 |
| ASB47762.1 | 1.53e-281 | 27 | 918 | 34 | 917 |
| APZ45922.1 | 1.31e-231 | 27 | 934 | 26 | 930 |
| AUC19784.1 | 1.31e-231 | 27 | 934 | 26 | 930 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4QVS_A | 1.42e-20 | 643 | 845 | 7 | 226 | ChainA, S-layer domain-containing protein [Acetivibrio thermocellus ATCC 27405] |
| 5HON_A | 1.61e-06 | 641 | 839 | 5 | 205 | Structureof Domain 4 of AbnA, a GH43 extracellular arabinanase from Geobacillus stearothermophilus, in complex with arabinotriose [Geobacillus stearothermophilus],5HON_B Structure of Domain 4 of AbnA, a GH43 extracellular arabinanase from Geobacillus stearothermophilus, in complex with arabinotriose [Geobacillus stearothermophilus] |
| 5HO0_A | 4.65e-06 | 622 | 839 | 608 | 844 | Crystalstructure of AbnA (closed conformation), a GH43 extracellular arabinanase from Geobacillus stearothermophilus [Geobacillus stearothermophilus],5HO2_A Crystal structure of AbnA (open conformation), a GH43 extracellular arabinanase from Geobacillus stearothermophilus [Geobacillus stearothermophilus],5HOF_A Crystal structure of AbnA, a GH43 extracellular arabinanase from Geobacillus stearothermophilus, in complex with arabinopentaose [Geobacillus stearothermophilus],5HP6_A Structure of AbnA, a GH43 extracellular arabinanase from Geobacillus stearothermophilus (a new conformational state) [Geobacillus stearothermophilus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
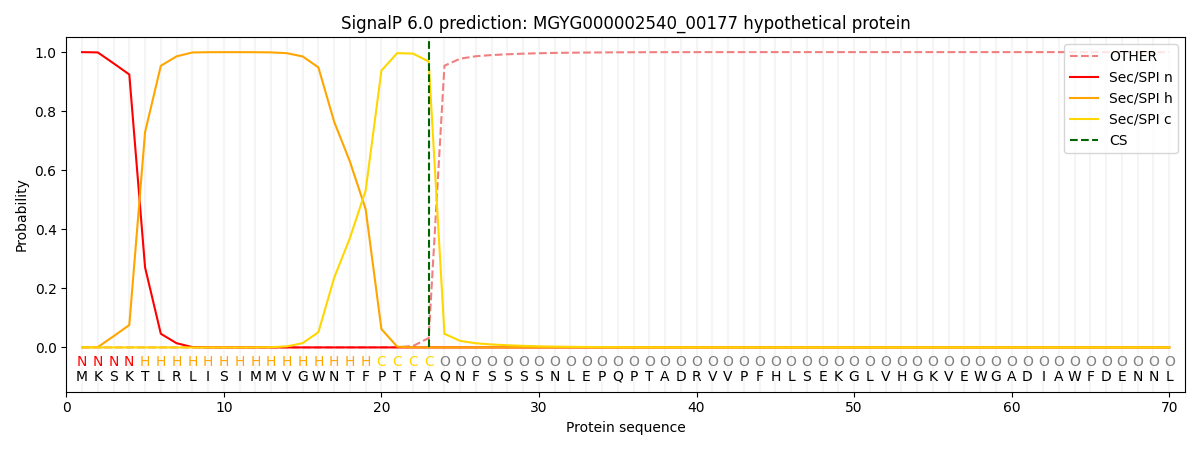
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000878 | 0.997671 | 0.000628 | 0.000308 | 0.000260 | 0.000233 |
