You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002540_01329
You are here: Home > Sequence: MGYG000002540_01329
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
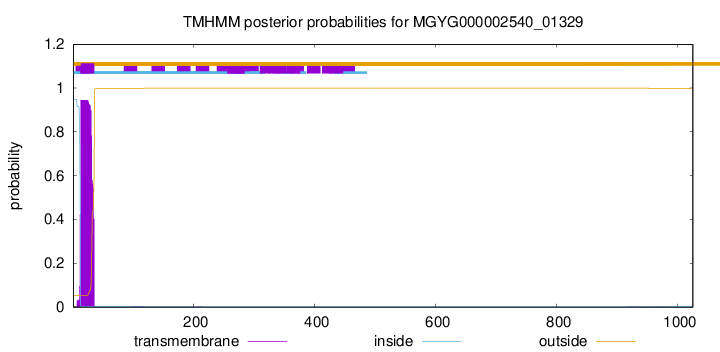
TMHMM annotations
Basic Information help
| Species | Paraprevotella clara | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Paraprevotella; Paraprevotella clara | |||||||||||
| CAZyme ID | MGYG000002540_01329 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 57760; End: 60837 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam05426 | Alginate_lyase | 7.26e-19 | 438 | 655 | 51 | 267 | Alginate lyase. This family contains several bacterial alginate lyase proteins. Alginate is a family of 1-4-linked copolymers of beta -D-mannuronic acid (M) and alpha -L-guluronic acid (G). It is produced by brown algae and by some bacteria belonging to the genera Azotobacter and Pseudomonas. Alginate lyases catalyze the depolymerization of alginates by beta -elimination, generating a molecule containing 4-deoxy-L-erythro-hex-4-enepyranosyluronate at the nonreducing end. This family adopts an all alpha fold. |
| pfam05426 | Alginate_lyase | 3.25e-16 | 64 | 296 | 15 | 267 | Alginate lyase. This family contains several bacterial alginate lyase proteins. Alginate is a family of 1-4-linked copolymers of beta -D-mannuronic acid (M) and alpha -L-guluronic acid (G). It is produced by brown algae and by some bacteria belonging to the genera Azotobacter and Pseudomonas. Alginate lyases catalyze the depolymerization of alginates by beta -elimination, generating a molecule containing 4-deoxy-L-erythro-hex-4-enepyranosyluronate at the nonreducing end. This family adopts an all alpha fold. |
| cd00063 | FN3 | 2.68e-11 | 746 | 835 | 1 | 92 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| smart00060 | FN3 | 1.40e-08 | 746 | 826 | 1 | 83 | Fibronectin type 3 domain. One of three types of internal repeat within the plasma protein, fibronectin. The tenth fibronectin type III repeat contains a RGD cell recognition sequence in a flexible loop between 2 strands. Type III modules are present in both extracellular and intracellular proteins. |
| COG3401 | FN3 | 0.001 | 746 | 836 | 257 | 343 | Fibronectin type 3 domain [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AXP82777.1 | 5.54e-206 | 381 | 1024 | 36 | 676 |
| QUT56879.1 | 4.14e-158 | 26 | 384 | 27 | 385 |
| QQY39805.1 | 1.59e-157 | 26 | 384 | 27 | 385 |
| ABR37862.1 | 1.59e-157 | 26 | 384 | 27 | 385 |
| AXP82778.1 | 2.09e-141 | 378 | 1024 | 24 | 675 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
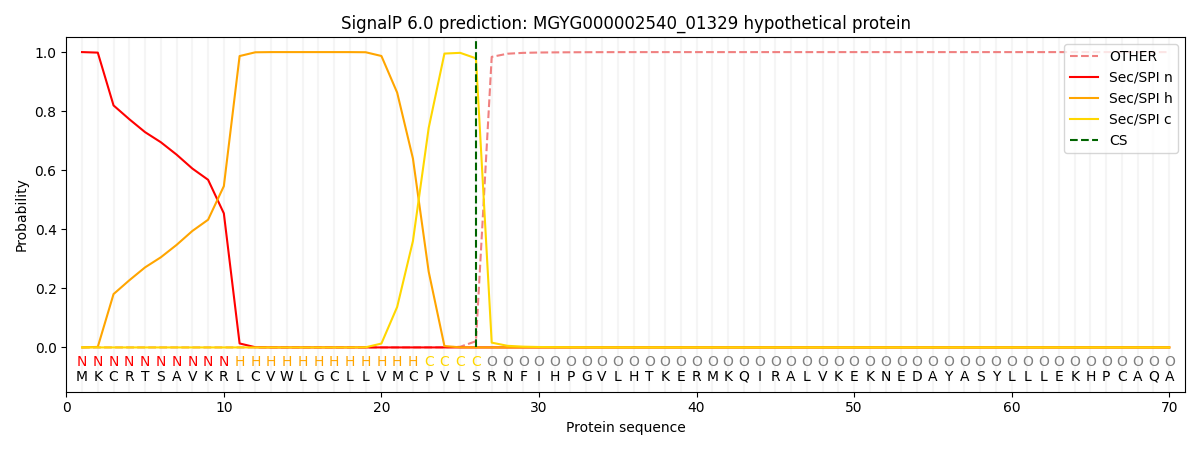
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000246 | 0.999186 | 0.000151 | 0.000143 | 0.000133 | 0.000128 |