You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002541_01234
You are here: Home > Sequence: MGYG000002541_01234
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
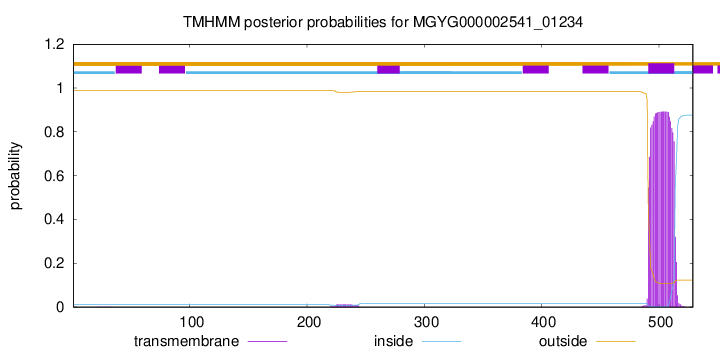
TMHMM annotations
Basic Information help
| Species | Cardiobacterium valvarum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Cardiobacteriales; Cardiobacteriaceae; Cardiobacterium; Cardiobacterium valvarum | |||||||||||
| CAZyme ID | MGYG000002541_01234 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20719; End: 22308 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 5 | 124 | 3.6e-16 | 0.7529411764705882 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd02511 | Beta4Glucosyltransferase | 2.02e-61 | 3 | 230 | 1 | 229 | UDP-glucose LOS-beta-1,4 glucosyltransferase is required for biosynthesis of lipooligosaccharide. UDP-glucose: lipooligosaccharide (LOS) beta-1-4-glucosyltransferase catalyzes the addition of the first residue, glucose, of the lacto-N-neotetrase structure to HepI of the LOS inner core. LOS is the major constituent of the outer leaflet of the outer membrane of gram-positive bacteria. It consists of a short oligosaccharide chain of variable composition (alpha chain) attached to a branched inner core which is lined in turn to lipid A. Beta 1,4 glucosyltransferase is required to attach the alpha chain to the inner core. |
| cd02511 | Beta4Glucosyltransferase | 2.47e-61 | 282 | 507 | 2 | 229 | UDP-glucose LOS-beta-1,4 glucosyltransferase is required for biosynthesis of lipooligosaccharide. UDP-glucose: lipooligosaccharide (LOS) beta-1-4-glucosyltransferase catalyzes the addition of the first residue, glucose, of the lacto-N-neotetrase structure to HepI of the LOS inner core. LOS is the major constituent of the outer leaflet of the outer membrane of gram-positive bacteria. It consists of a short oligosaccharide chain of variable composition (alpha chain) attached to a branched inner core which is lined in turn to lipid A. Beta 1,4 glucosyltransferase is required to attach the alpha chain to the inner core. |
| COG0463 | WcaA | 6.42e-19 | 1 | 244 | 2 | 254 | Glycosyltransferase involved in cell wall bisynthesis [Cell wall/membrane/envelope biogenesis]. |
| COG0463 | WcaA | 3.49e-16 | 282 | 376 | 5 | 107 | Glycosyltransferase involved in cell wall bisynthesis [Cell wall/membrane/envelope biogenesis]. |
| pfam00535 | Glycos_transf_2 | 1.94e-14 | 5 | 127 | 1 | 135 | Glycosyl transferase family 2. Diverse family, transferring sugar from UDP-glucose, UDP-N-acetyl- galactosamine, GDP-mannose or CDP-abequose, to a range of substrates including cellulose, dolichol phosphate and teichoic acids. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEG76449.1 | 9.14e-248 | 1 | 522 | 1 | 524 |
| ANB92440.1 | 8.58e-172 | 8 | 521 | 1 | 513 |
| AKG14536.1 | 6.94e-171 | 8 | 521 | 1 | 513 |
| AKG12579.1 | 6.94e-171 | 8 | 521 | 1 | 513 |
| AKG10553.1 | 6.94e-171 | 8 | 521 | 1 | 513 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9XC90 | 2.96e-55 | 282 | 521 | 5 | 245 | Lipopolysaccharide core biosynthesis glycosyltransferase WaaE OS=Klebsiella pneumoniae OX=573 GN=waaE PE=3 SV=1 |
| P44029 | 2.74e-46 | 4 | 249 | 4 | 251 | Uncharacterized glycosyltransferase HI_0653 OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=HI_0653 PE=3 SV=1 |
| Q54435 | 8.36e-38 | 4 | 244 | 7 | 247 | Lipopolysaccharide core biosynthesis glycosyltransferase KdtX OS=Serratia marcescens OX=615 GN=kdtX PE=3 SV=1 |
| O05944 | 7.91e-11 | 282 | 522 | 4 | 252 | Uncharacterized glycosyltransferase RP128 OS=Rickettsia prowazekii (strain Madrid E) OX=272947 GN=RP218 PE=3 SV=1 |
| P42460 | 1.25e-10 | 1 | 119 | 7 | 124 | TPR repeat-containing protein Synpcc7942_0270 OS=Synechococcus elongatus (strain PCC 7942 / FACHB-805) OX=1140 GN=Synpcc7942_0270 PE=4 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
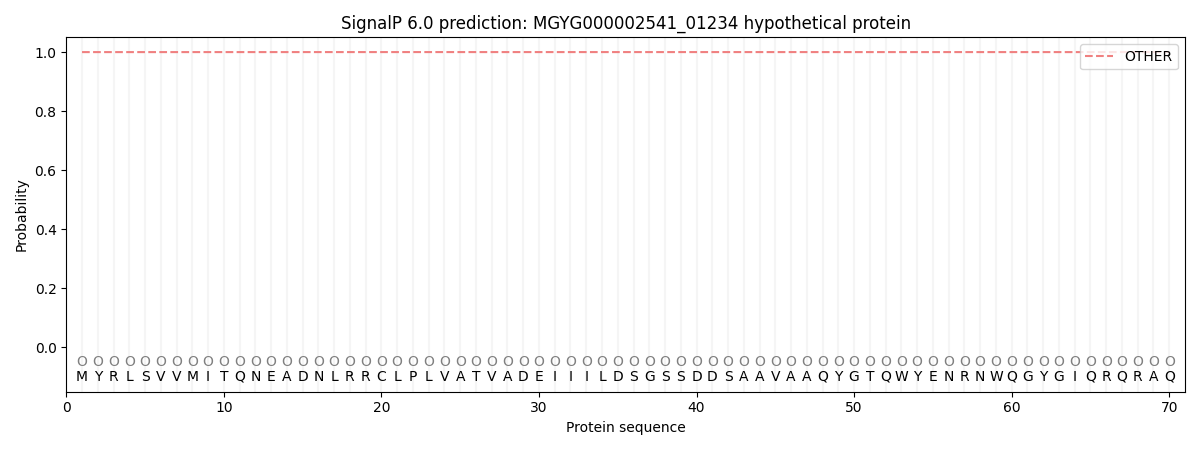
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000036 | 0.000009 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |