You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002542_01087
You are here: Home > Sequence: MGYG000002542_01087
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lentilactobacillus parafarraginis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Lentilactobacillus; Lentilactobacillus parafarraginis | |||||||||||
| CAZyme ID | MGYG000002542_01087 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1120; End: 3438 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 23 | 234 | 1.1e-29 | 0.387037037037037 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 1.22e-42 | 24 | 639 | 9 | 509 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231 | PMT_2 | 4.50e-33 | 78 | 232 | 1 | 154 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| COG4745 | COG4745 | 6.90e-06 | 92 | 237 | 78 | 227 | Predicted membrane-bound mannosyltransferase [General function prediction only]. |
| COG1928 | PMT1 | 1.53e-04 | 72 | 199 | 83 | 216 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| COG3127 | YbbP | 0.002 | 356 | 531 | 305 | 456 | Predicted ABC-type transport system involved in lysophospholipase L1 biosynthesis, permease component [Secondary metabolites biosynthesis, transport and catabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHU89858.1 | 1.79e-79 | 15 | 660 | 265 | 889 |
| QUB37647.1 | 1.91e-76 | 28 | 661 | 210 | 781 |
| QHU90800.1 | 6.84e-75 | 15 | 660 | 265 | 886 |
| QJU05422.1 | 7.39e-71 | 15 | 660 | 265 | 890 |
| QLF52252.1 | 6.87e-70 | 15 | 660 | 265 | 891 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34575 | 1.67e-180 | 15 | 673 | 2 | 677 | Putative mannosyltransferase YkcB OS=Bacillus subtilis (strain 168) OX=224308 GN=ykcB PE=3 SV=2 |
| P37483 | 3.96e-176 | 22 | 683 | 7 | 677 | Putative mannosyltransferase YycA OS=Bacillus subtilis (strain 168) OX=224308 GN=yycA PE=3 SV=2 |
| O67270 | 5.15e-06 | 24 | 235 | 8 | 213 | Uncharacterized protein aq_1220 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1220 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.862031 | 0.000607 | 0.000423 | 0.000003 | 0.000002 | 0.136954 |
TMHMM Annotations download full data without filtering help
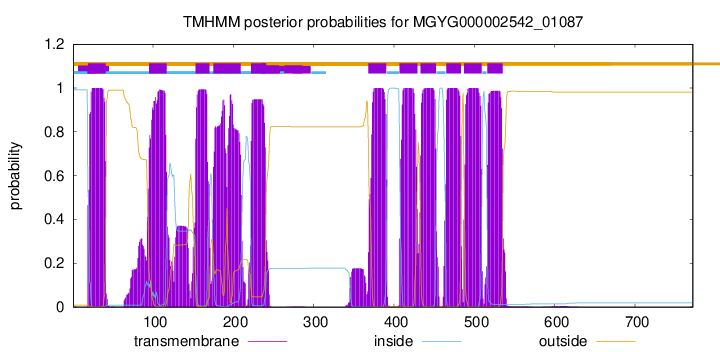
| start | end |
|---|---|
| 19 | 41 |
| 95 | 117 |
| 153 | 170 |
| 175 | 209 |
| 222 | 240 |
| 368 | 390 |
| 407 | 429 |
| 433 | 452 |
| 465 | 483 |
| 487 | 509 |
| 516 | 535 |
