You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002543_00572
You are here: Home > Sequence: MGYG000002543_00572
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
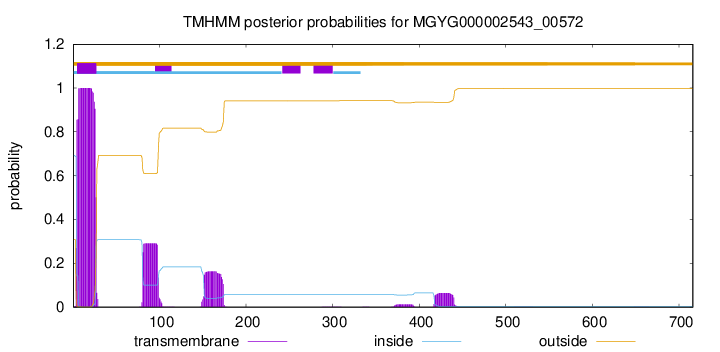
TMHMM annotations
Basic Information help
| Species | Campylobacter_B ureolyticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Campylobacterota; Campylobacteria; Campylobacterales; Campylobacteraceae; Campylobacter_B; Campylobacter_B ureolyticus | |||||||||||
| CAZyme ID | MGYG000002543_00572 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Monofunctional biosynthetic peptidoglycan transglycosylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 114179; End: 116329 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 61 | 217 | 2.4e-48 | 0.8983050847457628 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4953 | PbpC | 2.68e-144 | 5 | 576 | 4 | 583 | Membrane carboxypeptidase/penicillin-binding protein PbpC [Cell wall/membrane/envelope biogenesis]. |
| TIGR02073 | PBP_1c | 6.06e-142 | 38 | 558 | 11 | 547 | penicillin-binding protein 1C. This subfamily of the penicillin binding proteins includes the member from E. coli designated penicillin-binding protein 1C. Members have both transglycosylase and transpeptidase domains and are involved in forming cross-links in the late stages of peptidoglycan biosynthesis. All members of this subfamily are presumed to have the same basic function. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG0744 | MrcB | 2.87e-97 | 1 | 567 | 14 | 609 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| PRK11240 | PRK11240 | 7.28e-74 | 54 | 557 | 56 | 565 | penicillin-binding protein 1C; Provisional |
| COG5009 | MrcA | 1.40e-55 | 41 | 527 | 51 | 690 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIX85957.1 | 0.0 | 1 | 716 | 1 | 716 |
| QKF84157.1 | 0.0 | 1 | 716 | 1 | 716 |
| QQY35700.1 | 0.0 | 1 | 716 | 1 | 716 |
| AKT90485.1 | 0.0 | 1 | 716 | 1 | 716 |
| QKF64721.1 | 1.24e-310 | 2 | 711 | 5 | 712 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5FGZ_A | 3.86e-28 | 54 | 551 | 152 | 689 | E.coli PBP1b in complex with FPI-1465 [Escherichia coli K-12],5HL9_A E. coli PBP1b in complex with acyl-ampicillin and moenomycin [Escherichia coli K-12],5HLA_A E. coli PBP1b in complex with acyl-cephalexin and moenomycin [Escherichia coli K-12],5HLB_A E. coli PBP1b in complex with acyl-aztreonam and moenomycin [Escherichia coli K-12],5HLD_A E. coli PBP1b in complex with acyl-CENTA and moenomycin [Escherichia coli K-12],6YN0_A Structure of E. coli PBP1b with a FtsN peptide activating transglycosylase activity [Escherichia coli K-12],7LQ6_A Chain A, Penicillin-binding protein 1B [Escherichia coli K-12] |
| 3VMA_A | 4.05e-28 | 54 | 551 | 173 | 710 | CrystalStructure of the Full-Length Transglycosylase PBP1b from Escherichia coli [Escherichia coli K-12] |
| 3DWK_A | 7.74e-28 | 41 | 527 | 10 | 556 | ChainA, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_B Chain B, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_C Chain C, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_D Chain D, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL] |
| 3FWL_A | 3.71e-27 | 54 | 551 | 156 | 693 | CrystalStructure of the Full-Length Transglycosylase PBP1b from Escherichia coli [Escherichia coli] |
| 2OLU_A | 1.15e-25 | 41 | 527 | 19 | 565 | StructuralInsight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Apoenzyme [Staphylococcus aureus],2OLV_A Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Donor Ligand Complex [Staphylococcus aureus],2OLV_B Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Donor Ligand Complex [Staphylococcus aureus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P76577 | 9.43e-62 | 40 | 575 | 44 | 586 | Penicillin-binding protein 1C OS=Escherichia coli (strain K12) OX=83333 GN=pbpC PE=1 SV=1 |
| P40750 | 9.99e-38 | 29 | 527 | 46 | 596 | Penicillin-binding protein 4 OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpD PE=1 SV=2 |
| A7FY32 | 1.37e-33 | 37 | 528 | 55 | 618 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain ATCC 19397 / Type A) OX=441770 GN=pbpA PE=3 SV=1 |
| A5I6G4 | 1.37e-33 | 37 | 528 | 55 | 618 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Hall / ATCC 3502 / NCTC 13319 / Type A) OX=441771 GN=pbpA PE=3 SV=1 |
| Q89AR2 | 1.84e-33 | 60 | 560 | 159 | 697 | Penicillin-binding protein 1B OS=Buchnera aphidicola subsp. Baizongia pistaciae (strain Bp) OX=224915 GN=mrcB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
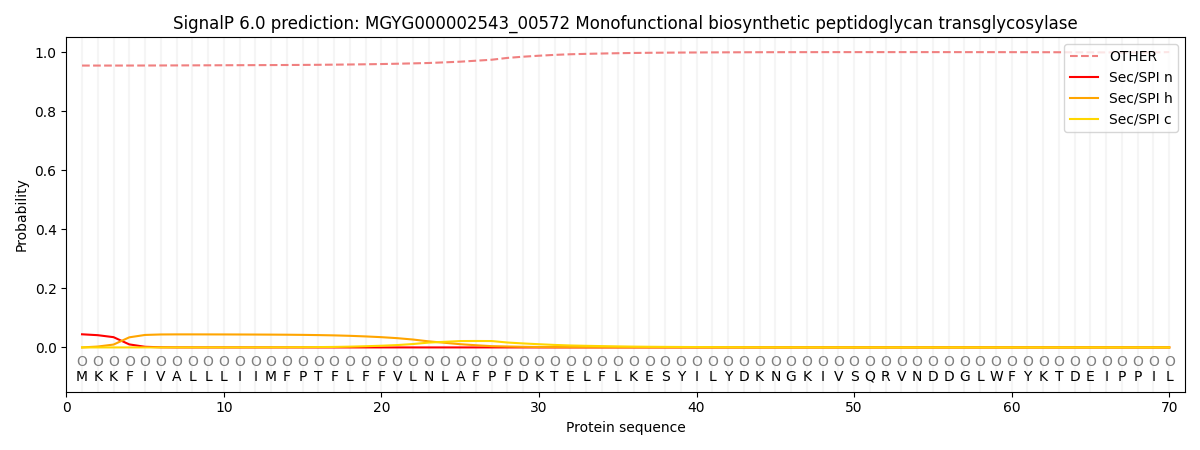
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.956673 | 0.041255 | 0.000732 | 0.000180 | 0.000128 | 0.001056 |