You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002548_01524
You are here: Home > Sequence: MGYG000002548_01524
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
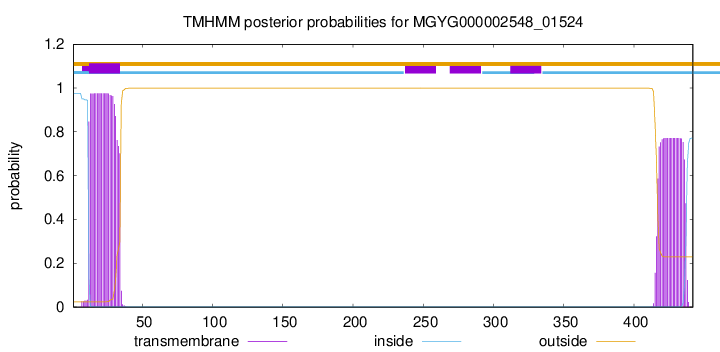
TMHMM annotations
Basic Information help
| Species | Staphylococcus argenteus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Staphylococcales; Staphylococcaceae; Staphylococcus; Staphylococcus argenteus | |||||||||||
| CAZyme ID | MGYG000002548_01524 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | Immunoglobulin G-binding protein A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 40607; End: 41935 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02216 | B | 1.32e-18 | 38 | 88 | 1 | 51 | B domain. This family contains the B domain of Staphylococcal protein A, which specifically binds to the Fc portion of immunoglobulin G. |
| pfam02216 | B | 3.99e-18 | 99 | 149 | 1 | 51 | B domain. This family contains the B domain of Staphylococcal protein A, which specifically binds to the Fc portion of immunoglobulin G. |
| pfam02216 | B | 5.87e-18 | 157 | 207 | 1 | 51 | B domain. This family contains the B domain of Staphylococcal protein A, which specifically binds to the Fc portion of immunoglobulin G. |
| pfam02216 | B | 9.33e-17 | 215 | 265 | 1 | 51 | B domain. This family contains the B domain of Staphylococcal protein A, which specifically binds to the Fc portion of immunoglobulin G. |
| cd00118 | LysM | 6.73e-13 | 348 | 391 | 2 | 45 | Lysin Motif is a small domain involved in binding peptidoglycan. LysM, a small globular domain with approximately 40 amino acids, is a widespread protein module involved in binding peptidoglycan in bacteria and chitin in eukaryotes. The domain was originally identified in enzymes that degrade bacterial cell walls, but proteins involved in many other biological functions also contain this domain. It has been reported that the LysM domain functions as a signal for specific plant-bacteria recognition in bacterial pathogenesis. Many of these enzymes are modular and are composed of catalytic units linked to one or several repeats of LysM domains. LysM domains are found in bacteria and eukaryotes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AXH79889.1 | 2.95e-220 | 1 | 442 | 1 | 442 |
| AXH79891.1 | 2.95e-220 | 1 | 442 | 1 | 442 |
| AXH79887.1 | 2.95e-220 | 1 | 442 | 1 | 442 |
| AXH79886.1 | 2.95e-220 | 1 | 442 | 1 | 442 |
| AXH79885.1 | 2.95e-220 | 1 | 442 | 1 | 442 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4NPF_X | 4.62e-65 | 154 | 267 | 1 | 114 | High-resolutionstructure of two tandem B domains of staphylococcal protein A connected by the conserved linker [Staphylococcus aureus],4NPF_Y High-resolution structure of two tandem B domains of staphylococcal protein A connected by the conserved linker [Staphylococcus aureus] |
| 4GWP_C | 9.13e-59 | 155 | 267 | 283 | 395 | Structureof the Mediator Head Module from S. cerevisiae [Saccharomyces cerevisiae S288C],4GWQ_C Structure of the Mediator Head Module from S. cerevisiae in complex with the carboxy-terminal domain (CTD) of RNA Polymerase II Rpb1 subunit [Saccharomyces cerevisiae S288C] |
| 7BG9_A | 3.57e-55 | 155 | 267 | 14 | 126 | ChainA, Telomerase reverse transcriptase,Telomerase reverse transcriptase [Homo sapiens] |
| 6WMP_D | 3.80e-55 | 155 | 267 | 1480 | 1592 | ChainD, DNA-directed RNA polymerase subunit beta' [Francisella tularensis subsp. holarctica LVS],6WMT_D Chain D, DNA-directed RNA polymerase subunit beta' [Francisella tularensis subsp. holarctica LVS] |
| 6HQA_A | 3.90e-55 | 155 | 267 | 1632 | 1744 | ChainA, Taf2 [Komagataella phaffii GS115] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0A015 | 9.17e-201 | 1 | 442 | 1 | 450 | Immunoglobulin G-binding protein A OS=Staphylococcus aureus (strain Mu50 / ATCC 700699) OX=158878 GN=spa PE=1 SV=1 |
| P99134 | 9.17e-201 | 1 | 442 | 1 | 450 | Immunoglobulin G-binding protein A OS=Staphylococcus aureus (strain N315) OX=158879 GN=spa PE=1 SV=1 |
| A0A0H3K686 | 1.35e-197 | 1 | 442 | 1 | 508 | Immunoglobulin G-binding protein A OS=Staphylococcus aureus (strain Newman) OX=426430 GN=spa PE=1 SV=2 |
| P38507 | 1.10e-196 | 1 | 442 | 1 | 508 | Immunoglobulin G-binding protein A OS=Staphylococcus aureus OX=1280 GN=spa PE=1 SV=1 |
| P02976 | 2.92e-196 | 1 | 442 | 1 | 516 | Immunoglobulin G-binding protein A OS=Staphylococcus aureus (strain NCTC 8325 / PS 47) OX=93061 GN=spa PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000265 | 0.999020 | 0.000172 | 0.000205 | 0.000164 | 0.000139 |