You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002558_01727
You are here: Home > Sequence: MGYG000002558_01727
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900540415 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900540415 | |||||||||||
| CAZyme ID | MGYG000002558_01727 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6966; End: 8720 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 325 | 552 | 1.1e-27 | 0.5907590759075908 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 3.00e-15 | 324 | 533 | 51 | 222 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.13e-06 | 331 | 497 | 123 | 260 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRQ58664.1 | 1.46e-134 | 3 | 576 | 5 | 582 |
| SCV06584.1 | 1.46e-134 | 3 | 576 | 5 | 582 |
| ALJ47172.1 | 1.46e-134 | 3 | 576 | 5 | 582 |
| QUT25655.1 | 1.46e-134 | 3 | 576 | 5 | 582 |
| ALJ47155.1 | 1.06e-104 | 41 | 573 | 132 | 684 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1US3_A | 3.32e-13 | 302 | 550 | 254 | 472 | Nativexylanase10C from Cellvibrio japonicus [Cellvibrio japonicus] |
| 1US2_A | 1.79e-12 | 302 | 550 | 254 | 472 | Xylanase10C(mutant E385A) from Cellvibrio japonicus in complex with xylopentaose [Cellvibrio japonicus] |
| 1TUX_A | 8.17e-10 | 328 | 497 | 101 | 240 | HighResolution Crystal Structure Of A Thermostable Xylanase From Thermoascus Aurantiacus [Thermoascus aurantiacus] |
| 2Q8X_A | 1.79e-08 | 313 | 556 | 85 | 311 | Thehigh-resolution crystal structure of ixt6, a thermophilic, intracellular xylanase from G. stearothermophilus [unidentified],2Q8X_B The high-resolution crystal structure of ixt6, a thermophilic, intracellular xylanase from G. stearothermophilus [unidentified],3MSD_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSD_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSG_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSG_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus] |
| 1N82_A | 3.18e-08 | 313 | 556 | 85 | 311 | Thehigh-resolution crystal structure of IXT6, a thermophilic, intracellular xylanase from G. stearothermophilus [Geobacillus stearothermophilus],1N82_B The high-resolution crystal structure of IXT6, a thermophilic, intracellular xylanase from G. stearothermophilus [Geobacillus stearothermophilus],3MUA_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MUA_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q59675 | 2.09e-12 | 302 | 550 | 338 | 556 | Endo-beta-1,4-xylanase Xyn10C OS=Cellvibrio japonicus OX=155077 GN=xyn10C PE=1 SV=2 |
| Q6PRW6 | 2.67e-09 | 328 | 497 | 131 | 270 | Endo-1,4-beta-xylanase OS=Penicillium chrysogenum OX=5076 GN=Xyn PE=1 SV=1 |
| W0HFK8 | 3.12e-09 | 328 | 497 | 131 | 270 | Endo-1,4-beta-xylanase 1 OS=Rhizopus oryzae OX=64495 GN=xyn1 PE=1 SV=1 |
| P29417 | 3.56e-09 | 328 | 497 | 131 | 270 | Endo-1,4-beta-xylanase OS=Penicillium chrysogenum OX=5076 GN=XYLP PE=1 SV=2 |
| G4MTF8 | 5.54e-08 | 328 | 497 | 129 | 269 | Endo-1,4-beta-xylanase 2 OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=XYL2 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
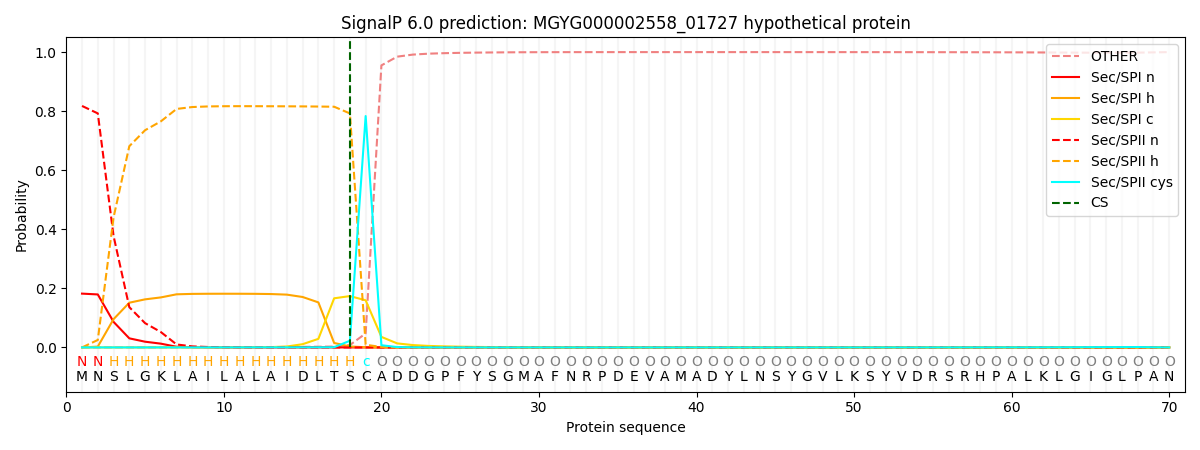
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000635 | 0.177246 | 0.821783 | 0.000111 | 0.000119 | 0.000114 |
