You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002560_00609
You are here: Home > Sequence: MGYG000002560_00609
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
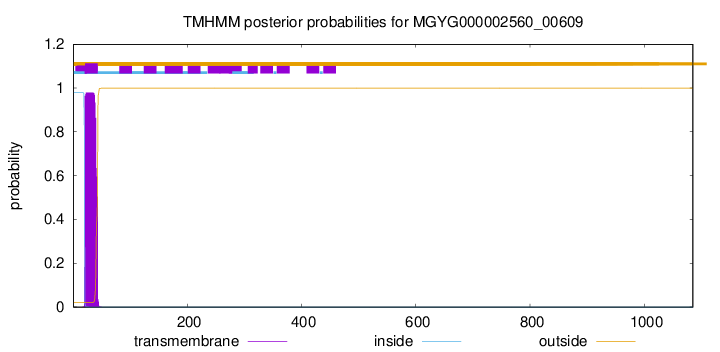
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp902388365 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp902388365 | |||||||||||
| CAZyme ID | MGYG000002560_00609 | |||||||||||
| CAZy Family | GH8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 200795; End: 204052 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH67 | 449 | 1053 | 6.3e-270 | 0.9327354260089686 |
| GH8 | 100 | 426 | 9.3e-80 | 0.928125 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3661 | AguA2 | 0.0 | 507 | 1054 | 108 | 677 | Alpha-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam07488 | Glyco_hydro_67M | 0.0 | 528 | 828 | 1 | 324 | Glycosyl hydrolase family 67 middle domain. Alpha-glucuronidases, components of an ensemble of enzymes central to the recycling of photosynthetic biomass, remove the alpha-1,2 linked 4-O-methyl glucuronic acid from xylans. This family represents the central catalytic domain of alpha-glucuronidase. |
| pfam07477 | Glyco_hydro_67C | 2.93e-132 | 830 | 1057 | 1 | 223 | Glycosyl hydrolase family 67 C-terminus. Alpha-glucuronidases, components of an ensemble of enzymes central to the recycling of photosynthetic biomass, remove the alpha-1,2 linked 4-O-methyl glucuronic acid from xylans. This family represents the C terminal region of alpha-glucuronidase which is mainly alpha-helical. It wraps around the catalytic domain (pfam07488), making additional interactions both with the N-terminal domain (pfam03648) of its parent monomer and also forming the majority of the dimer-surface with the equivalent C-terminal domain of the other monomer of the dimer. |
| COG3405 | BcsZ | 1.36e-45 | 92 | 439 | 37 | 357 | Endo-1,4-beta-D-glucanase Y [Carbohydrate transport and metabolism]. |
| pfam01270 | Glyco_hydro_8 | 1.03e-20 | 102 | 320 | 28 | 233 | Glycosyl hydrolases family 8. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQY39903.1 | 0.0 | 440 | 1085 | 34 | 681 |
| ABR37759.1 | 0.0 | 440 | 1085 | 34 | 681 |
| ALK86291.1 | 0.0 | 440 | 1085 | 34 | 681 |
| QUT56985.1 | 0.0 | 440 | 1085 | 34 | 681 |
| QJR73691.1 | 0.0 | 441 | 1085 | 35 | 681 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1GQI_A | 1.05e-227 | 442 | 1067 | 1 | 686 | Structureof Pseudomonas cellulosa alpha-D-glucuronidase [Cellvibrio japonicus],1GQI_B Structure of Pseudomonas cellulosa alpha-D-glucuronidase [Cellvibrio japonicus],1GQJ_A Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with xylobiose [Cellvibrio japonicus],1GQJ_B Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with xylobiose [Cellvibrio japonicus],1GQK_A Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with glucuronic acid [Cellvibrio japonicus],1GQK_B Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with glucuronic acid [Cellvibrio japonicus],1GQL_A Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with glucuronic acid and xylotriose [Cellvibrio japonicus],1GQL_B Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with glucuronic acid and xylotriose [Cellvibrio japonicus] |
| 1H41_A | 8.32e-227 | 442 | 1067 | 1 | 686 | Pseudomonascellulosa E292A alpha-D-glucuronidase mutant complexed with aldotriuronic acid [Cellvibrio japonicus],1H41_B Pseudomonas cellulosa E292A alpha-D-glucuronidase mutant complexed with aldotriuronic acid [Cellvibrio japonicus] |
| 1MQP_A | 2.28e-171 | 508 | 1054 | 104 | 673 | TheCrystal Structure Of Alpha-D-Glucuronidase From Bacillus Stearothermophilus T-6 [Geobacillus stearothermophilus] |
| 1K9D_A | 4.52e-171 | 508 | 1054 | 104 | 673 | The1.7 A crystal structure of alpha-D-glucuronidase, a family-67 glycoside hydrolase from Bacillus stearothermophilus T-1 [Geobacillus stearothermophilus],1L8N_A The 1.5A crystal structure of alpha-D-glucuronidase from Bacillus stearothermophilus T-1, complexed with 4-O-methyl-glucuronic acid and xylotriose [Geobacillus stearothermophilus],1MQQ_A THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE FROM BACILLUS STEAROTHERMOPHILUS T-1 COMPLEXED WITH GLUCURONIC ACID [Geobacillus stearothermophilus] |
| 1MQR_A | 6.36e-171 | 508 | 1054 | 104 | 673 | ChainA, ALPHA-D-GLUCURONIDASE [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B3PC73 | 2.30e-227 | 436 | 1067 | 19 | 710 | Extracellular xylan exo-alpha-(1->2)-glucuronosidase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=gla67A PE=1 SV=1 |
| Q09LY5 | 1.25e-170 | 508 | 1054 | 104 | 673 | Xylan alpha-(1->2)-glucuronosidase OS=Geobacillus stearothermophilus OX=1422 GN=aguA PE=1 SV=1 |
| P96105 | 4.61e-165 | 510 | 1051 | 103 | 665 | Xylan alpha-(1->2)-glucuronosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=aguA PE=1 SV=2 |
| Q5AQZ4 | 1.36e-138 | 497 | 1079 | 113 | 728 | Alpha-glucuronidase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=aguA PE=1 SV=1 |
| A1CC12 | 4.32e-138 | 495 | 1067 | 111 | 706 | Probable alpha-glucuronidase A OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=aguA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
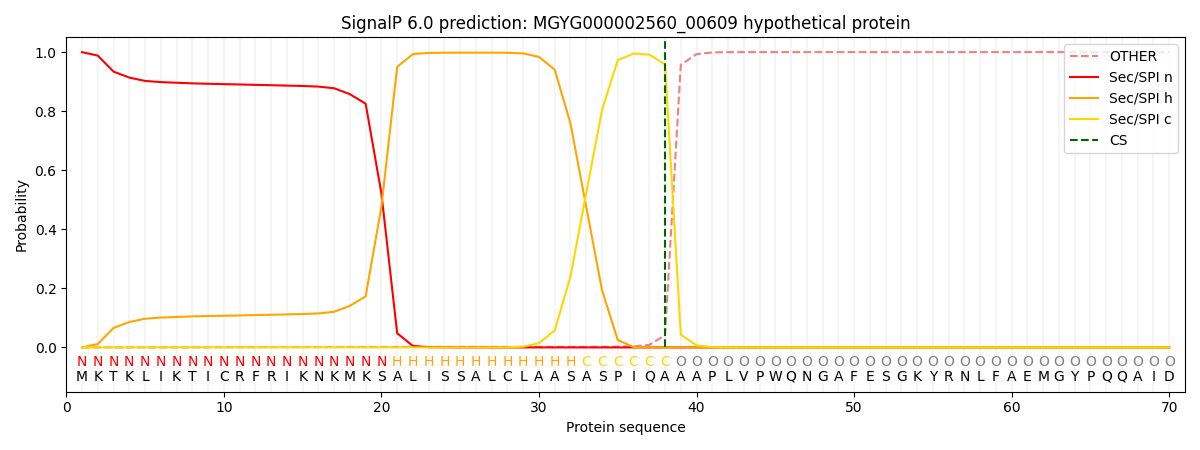
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000565 | 0.998488 | 0.000335 | 0.000237 | 0.000186 | 0.000160 |