You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002561_02719
You are here: Home > Sequence: MGYG000002561_02719
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp902388495 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp902388495 | |||||||||||
| CAZyme ID | MGYG000002561_02719 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 107; End: 1483 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 74 | 257 | 5.8e-77 | 0.9890710382513661 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3866 | PelB | 1.19e-07 | 4 | 257 | 11 | 273 | Pectate lyase [Carbohydrate transport and metabolism]. |
| smart00656 | Amb_all | 8.85e-05 | 73 | 257 | 1 | 186 | Amb_all domain. |
| pfam18884 | TSP3_bac | 7.00e-04 | 413 | 433 | 1 | 21 | Bacterial TSP3 repeat. This entry contains a novel bacterial thrombospondin type 3 repeat which differs from the typical consensus by containing a glutamate in place of one of the calcium binding aspartate residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ59319.1 | 8.96e-295 | 1 | 458 | 1 | 458 |
| QCD40703.1 | 8.23e-246 | 13 | 458 | 8 | 454 |
| QCP73753.1 | 8.23e-246 | 13 | 458 | 8 | 454 |
| ADY37134.1 | 1.55e-214 | 1 | 458 | 1 | 431 |
| ADY54248.1 | 1.16e-172 | 9 | 457 | 16 | 442 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FI2_A | 5.64e-14 | 28 | 238 | 46 | 253 | VexL:A periplasmic depolymerase provides new insight into ABC transporter-dependent secretion of bacterial capsular polysaccharides [Achromobacter denitrificans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8NQQ7 | 7.98e-56 | 22 | 457 | 21 | 414 | Probable pectate lyase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyC PE=3 SV=1 |
| Q5B297 | 1.64e-55 | 22 | 457 | 21 | 411 | Probable pectate lyase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyC PE=3 SV=1 |
| Q0CLG7 | 1.17e-54 | 11 | 457 | 10 | 414 | Probable pectate lyase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyC PE=3 SV=1 |
| Q2UB83 | 4.48e-54 | 22 | 457 | 21 | 414 | Probable pectate lyase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=plyC PE=3 SV=1 |
| Q4WL88 | 1.30e-52 | 10 | 457 | 8 | 415 | Probable pectate lyase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
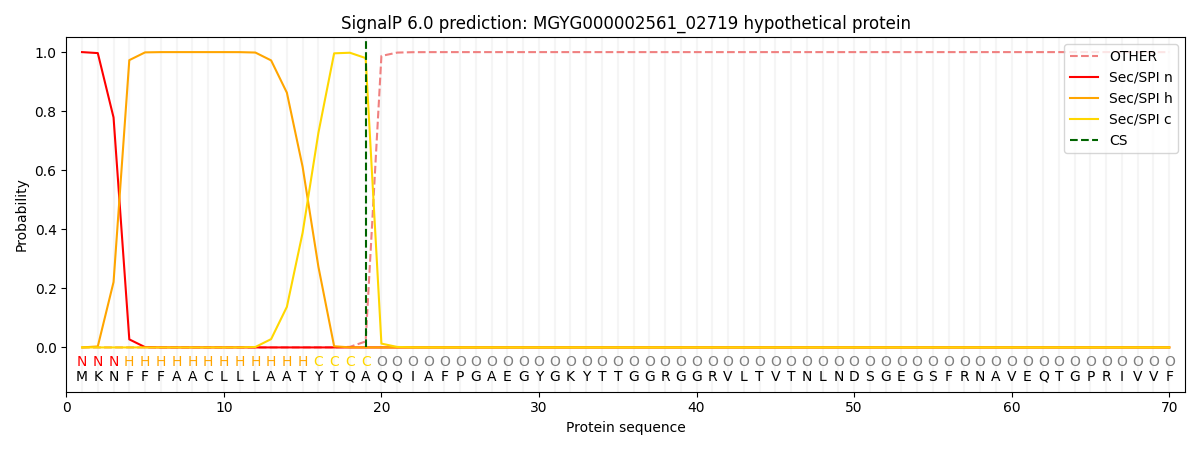
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000319 | 0.998925 | 0.000193 | 0.000198 | 0.000193 | 0.000167 |
