You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002563_00634
You are here: Home > Sequence: MGYG000002563_00634
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Romboutsia timonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Peptostreptococcales; Peptostreptococcaceae; Romboutsia; Romboutsia timonensis | |||||||||||
| CAZyme ID | MGYG000002563_00634 | |||||||||||
| CAZy Family | SLH | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 36221; End: 38395 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| SLH | 610 | 651 | 3.6e-16 | 0.9761904761904762 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 1.43e-10 | 301 | 458 | 78 | 244 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| smart00047 | LYZ2 | 1.96e-09 | 319 | 448 | 5 | 145 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| pfam00395 | SLH | 2.11e-09 | 610 | 651 | 1 | 42 | S-layer homology domain. |
| pfam01832 | Glucosaminidase | 7.02e-09 | 327 | 390 | 1 | 75 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| pfam00395 | SLH | 5.55e-08 | 672 | 715 | 1 | 42 | S-layer homology domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CED94675.1 | 0.0 | 12 | 724 | 17 | 729 |
| CEI73319.1 | 6.43e-151 | 23 | 548 | 22 | 534 |
| AXH51676.1 | 1.11e-111 | 48 | 579 | 72 | 614 |
| AWS25220.1 | 1.56e-111 | 48 | 549 | 72 | 579 |
| ASY50729.1 | 1.56e-111 | 48 | 549 | 72 | 579 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4Q2W_A | 1.27e-15 | 302 | 447 | 147 | 287 | CrystalStructure of pneumococcal peptidoglycan hydrolase LytB [Streptococcus pneumoniae TIGR4] |
| 6BT4_A | 2.83e-11 | 608 | 722 | 24 | 133 | Crystalstructure of the SLH domain of Sap from Bacillus anthracis in complex with a pyruvylated SCWP unit [Bacillus anthracis] |
| 3PYW_A | 2.89e-11 | 608 | 722 | 3 | 112 | Thestructure of the SLH domain from B. anthracis surface array protein at 1.8A [Bacillus anthracis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38536 | 5.89e-18 | 556 | 719 | 1684 | 1851 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P38535 | 1.45e-17 | 556 | 719 | 910 | 1077 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| P38537 | 5.35e-16 | 562 | 721 | 47 | 204 | Surface-layer 125 kDa protein OS=Lysinibacillus sphaericus OX=1421 PE=3 SV=1 |
| C6CRV0 | 2.28e-15 | 561 | 721 | 1291 | 1457 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
| P59205 | 9.92e-15 | 302 | 447 | 515 | 655 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=lytB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
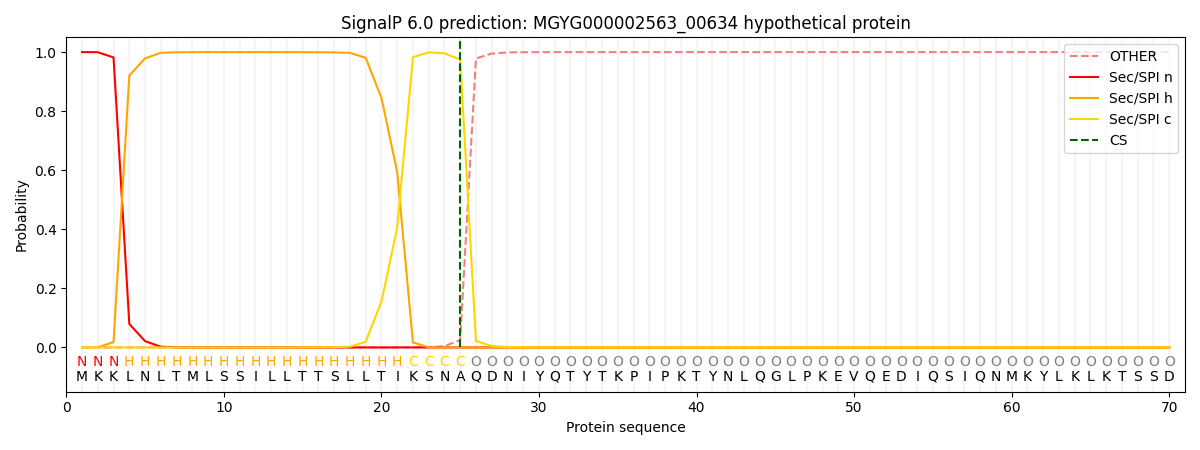
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000284 | 0.998941 | 0.000274 | 0.000173 | 0.000157 | 0.000141 |
