You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002563_02153
You are here: Home > Sequence: MGYG000002563_02153
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
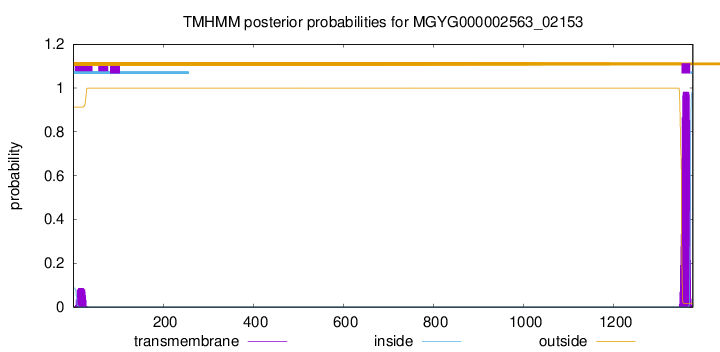
TMHMM annotations
Basic Information help
| Species | Romboutsia timonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Peptostreptococcales; Peptostreptococcaceae; Romboutsia; Romboutsia timonensis | |||||||||||
| CAZyme ID | MGYG000002563_02153 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | Trifunctional nucleotide phosphoesterase protein YfkN | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7534; End: 11664 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK09419 | PRK09419 | 0.0 | 106 | 1216 | 33 | 1143 | multifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase/5'-nucleotidase. |
| PRK09420 | cpdB | 0.0 | 106 | 692 | 17 | 606 | bifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase. |
| PRK11907 | PRK11907 | 9.81e-180 | 112 | 739 | 113 | 738 | bifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase. |
| PRK09418 | PRK09418 | 2.63e-160 | 107 | 736 | 32 | 660 | bifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase. |
| COG0737 | UshA | 2.31e-97 | 739 | 1210 | 26 | 503 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/5'- or 3'-nucleotidase, 5'-nucleotidase family [Nucleotide transport and metabolism, Defense mechanisms]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VDN48620.1 | 9.45e-147 | 100 | 1207 | 76 | 1118 |
| QUI23903.1 | 1.56e-146 | 103 | 1230 | 71 | 1137 |
| QQY79959.1 | 2.64e-145 | 733 | 1262 | 32 | 554 |
| QUH21387.1 | 1.53e-143 | 739 | 1217 | 38 | 508 |
| QZY54554.1 | 1.91e-143 | 727 | 1236 | 25 | 516 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EQV_A | 1.08e-59 | 106 | 396 | 1 | 289 | 1.45Angstrom Crystal Structure of Bifunctional 2',3'-cyclic Nucleotide 2'-phosphodiesterase/3'-Nucleotidase Periplasmic Precursor Protein from Yersinia pestis with Phosphate bound to the Active site [Yersinia pestis CO92] |
| 3JYF_A | 6.64e-57 | 111 | 396 | 5 | 288 | ChainA, 2',3'-cyclic nucleotide 2'-phosphodiesterase/3'-nucleotidase bifunctional periplasmic protein [Klebsiella pneumoniae subsp. pneumoniae MGH 78578],3JYF_B Chain B, 2',3'-cyclic nucleotide 2'-phosphodiesterase/3'-nucleotidase bifunctional periplasmic protein [Klebsiella pneumoniae subsp. pneumoniae MGH 78578] |
| 3GVE_A | 2.05e-55 | 106 | 444 | 3 | 339 | Crystalstructure of calcineurin-like phosphoesterase YfkN from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3GVE_B Crystal structure of calcineurin-like phosphoesterase YfkN from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168] |
| 2Z1A_A | 1.38e-52 | 740 | 1216 | 30 | 528 | Crystalstructure of 5'-nucleotidase precursor from Thermus thermophilus HB8 [Thermus thermophilus HB8] |
| 7D0V_A | 7.29e-52 | 739 | 1221 | 4 | 529 | ChainA, Snake venom 5'-nucleotidase [Naja atra],7D0V_B Chain B, Snake venom 5'-nucleotidase [Naja atra] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34313 | 1.74e-218 | 103 | 1216 | 33 | 1174 | Trifunctional nucleotide phosphoesterase protein YfkN OS=Bacillus subtilis (strain 168) OX=224308 GN=yfkN PE=1 SV=1 |
| P08331 | 6.03e-136 | 111 | 686 | 20 | 598 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase OS=Escherichia coli (strain K12) OX=83333 GN=cpdB PE=1 SV=2 |
| P44764 | 1.67e-131 | 110 | 723 | 29 | 652 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=cpdB PE=3 SV=1 |
| P26265 | 6.59e-129 | 111 | 688 | 20 | 600 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=cpdB PE=3 SV=2 |
| P53052 | 1.49e-126 | 102 | 688 | 16 | 605 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase OS=Yersinia enterocolitica OX=630 GN=cpdB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
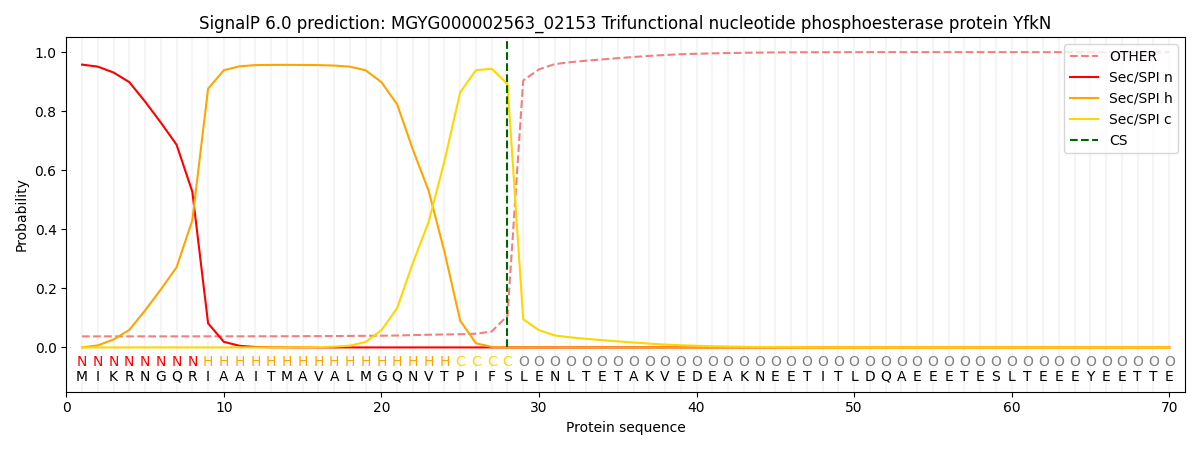
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.044187 | 0.948927 | 0.005939 | 0.000363 | 0.000287 | 0.000264 |