You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002575_00293
You are here: Home > Sequence: MGYG000002575_00293
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; ; | |||||||||||
| CAZyme ID | MGYG000002575_00293 | |||||||||||
| CAZy Family | GH128 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12095; End: 15409 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH128 | 244 | 479 | 2.7e-34 | 0.9642857142857143 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam11790 | Glyco_hydro_cc | 5.25e-21 | 232 | 480 | 6 | 235 | Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781. |
| pfam13385 | Laminin_G_3 | 2.97e-17 | 838 | 1001 | 5 | 151 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| COG3401 | FN3 | 1.32e-09 | 532 | 780 | 101 | 341 | Fibronectin type 3 domain [General function prediction only]. |
| TIGR04183 | Por_Secre_tail | 3.18e-09 | 1028 | 1104 | 1 | 72 | Por secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber average twenty or more copies of a C-terminal domain, represented by this model, associated with sorting to the outer membrane and covalent modification. |
| cd00063 | FN3 | 9.97e-09 | 601 | 686 | 3 | 93 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUC19783.1 | 0.0 | 50 | 1104 | 365 | 1414 |
| APZ45921.1 | 0.0 | 50 | 1104 | 365 | 1414 |
| AXY72824.1 | 1.49e-251 | 55 | 1078 | 395 | 1386 |
| AEV99551.1 | 4.06e-235 | 58 | 1008 | 419 | 1341 |
| QGK74225.1 | 1.18e-142 | 54 | 797 | 555 | 1297 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6UAQ_A | 3.18e-16 | 245 | 387 | 43 | 194 | Crystalstructure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) [Amycolatopsis mediterranei],6UAR_A Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaritriose [Amycolatopsis mediterranei] |
| 6UFL_A | 7.84e-16 | 245 | 387 | 43 | 194 | Crystalstructure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199Q mutant) from Amycolatopsis mediterranei (AmGH128_I) in the complex with laminarihexaose [Amycolatopsis mediterranei],6UFZ_A Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199Q mutant) from Amycolatopsis mediterranei (AmGH128_I) [Amycolatopsis mediterranei] |
| 6UAS_A | 1.93e-15 | 245 | 385 | 43 | 192 | Crystalstructure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199A mutant) from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaripentaose [Amycolatopsis mediterranei] |
| 6UAT_A | 1.93e-15 | 245 | 387 | 43 | 194 | Crystalstructure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E102A mutant) from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaripentaose [Amycolatopsis mediterranei],6UAU_A Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E102A mutant) from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaritriose and laminaribiose [Amycolatopsis mediterranei] |
| 3MPC_A | 5.05e-10 | 599 | 680 | 2 | 87 | ChainA, Fn3-like protein [Acetivibrio thermocellus ATCC 27405],3MPC_B Chain B, Fn3-like protein [Acetivibrio thermocellus ATCC 27405] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q09788 | 2.14e-06 | 232 | 407 | 294 | 473 | Alkali-sensitive linkage protein 1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=asl1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
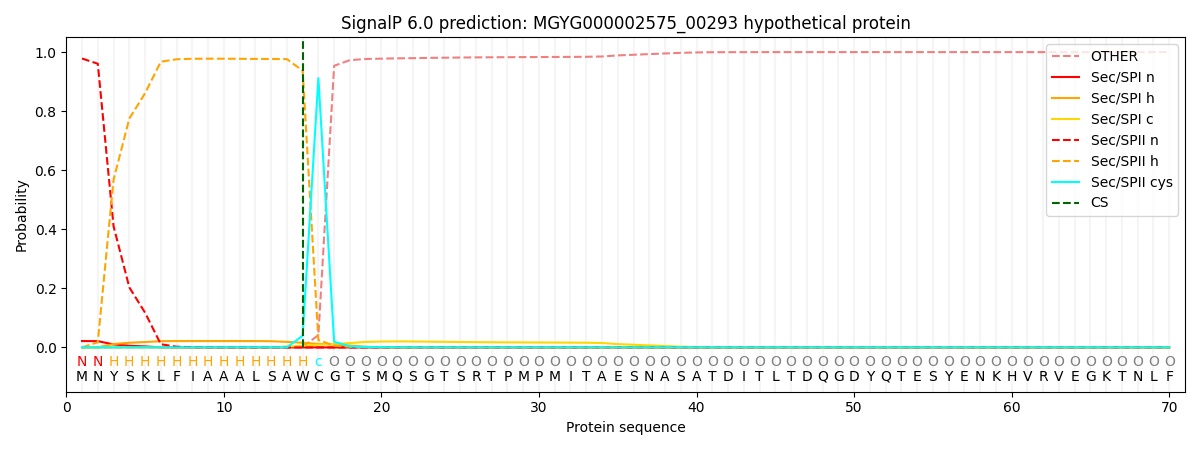
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000216 | 0.021032 | 0.978735 | 0.000009 | 0.000013 | 0.000009 |
