You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002580_00197
You are here: Home > Sequence: MGYG000002580_00197
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
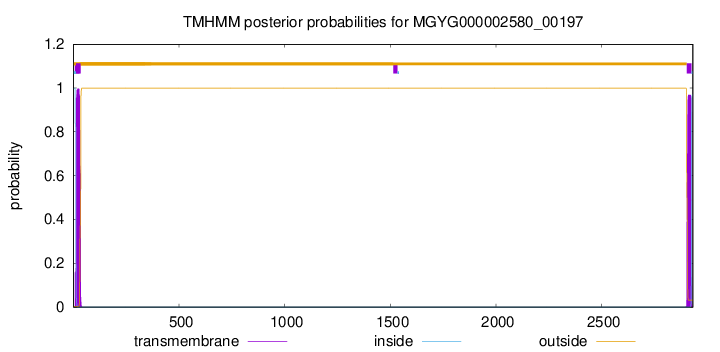
TMHMM annotations
Basic Information help
| Species | Lachnospira sp000436475 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnospira; Lachnospira sp000436475 | |||||||||||
| CAZyme ID | MGYG000002580_00197 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 201571; End: 210369 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 1275 | 1556 | 1e-75 | 0.9513888888888888 |
| PL1 | 1707 | 1944 | 2e-40 | 0.801980198019802 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 2.88e-38 | 1258 | 1553 | 81 | 395 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02432 | PLN02432 | 3.57e-35 | 1279 | 1553 | 23 | 275 | putative pectinesterase |
| PLN02773 | PLN02773 | 1.82e-34 | 1258 | 1536 | 4 | 269 | pectinesterase |
| pfam01095 | Pectinesterase | 7.58e-32 | 1258 | 1536 | 1 | 265 | Pectinesterase. |
| smart00656 | Amb_all | 1.46e-30 | 1708 | 1918 | 15 | 171 | Amb_all domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADD62022.1 | 0.0 | 19 | 2102 | 1 | 2087 |
| QEH70392.1 | 0.0 | 763 | 2097 | 489 | 1794 |
| ACZ98653.1 | 0.0 | 685 | 2049 | 3 | 1353 |
| ACR72234.1 | 7.70e-133 | 781 | 1571 | 278 | 1077 |
| QJS18636.1 | 1.87e-78 | 1609 | 2044 | 244 | 673 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5GT5_A | 2.55e-69 | 1610 | 2044 | 7 | 446 | Structuralbasis of the specific activity and thermostability of pectate lyase (pelN) from Paenibacillus sp. 0602 [Paenibacillus sp. 0602],5GT5_B Structural basis of the specific activity and thermostability of pectate lyase (pelN) from Paenibacillus sp. 0602 [Paenibacillus sp. 0602] |
| 2NSP_A | 1.09e-28 | 1255 | 1564 | 2 | 339 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
| 3UW0_A | 2.23e-25 | 1242 | 1550 | 16 | 348 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
| 1QJV_A | 2.72e-25 | 1255 | 1564 | 2 | 339 | ChainA, PECTIN METHYLESTERASE [Dickeya chrysanthemi],1QJV_B Chain B, PECTIN METHYLESTERASE [Dickeya chrysanthemi] |
| 2NTB_A | 9.03e-25 | 1255 | 1564 | 2 | 339 | Crystalstructure of pectin methylesterase in complex with hexasaccharide V [Dickeya dadantii 3937],2NTB_B Crystal structure of pectin methylesterase in complex with hexasaccharide V [Dickeya dadantii 3937],2NTP_A Crystal structure of pectin methylesterase in complex with hexasaccharide VI [Dickeya dadantii 3937],2NTP_B Crystal structure of pectin methylesterase in complex with hexasaccharide VI [Dickeya dadantii 3937],2NTQ_A Crystal structure of pectin methylesterase in complex with hexasaccharide VII [Dickeya dadantii 3937],2NTQ_B Crystal structure of pectin methylesterase in complex with hexasaccharide VII [Dickeya dadantii 3937] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D3JTC2 | 3.97e-67 | 1610 | 1941 | 36 | 366 | Pectate lyase B OS=Paenibacillus amylolyticus OX=1451 GN=pelB PE=1 SV=1 |
| P0C1A8 | 3.09e-24 | 1255 | 1564 | 26 | 363 | Pectinesterase A OS=Dickeya chrysanthemi OX=556 GN=pemA PE=1 SV=1 |
| Q9ZQA3 | 3.28e-24 | 1273 | 1565 | 95 | 384 | Probable pectinesterase 15 OS=Arabidopsis thaliana OX=3702 GN=PME15 PE=2 SV=1 |
| Q9FM79 | 3.90e-24 | 1229 | 1506 | 50 | 317 | Pectinesterase QRT1 OS=Arabidopsis thaliana OX=3702 GN=QRT1 PE=1 SV=1 |
| Q9LVQ0 | 7.55e-24 | 1274 | 1553 | 12 | 286 | Pectinesterase 31 OS=Arabidopsis thaliana OX=3702 GN=PME31 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
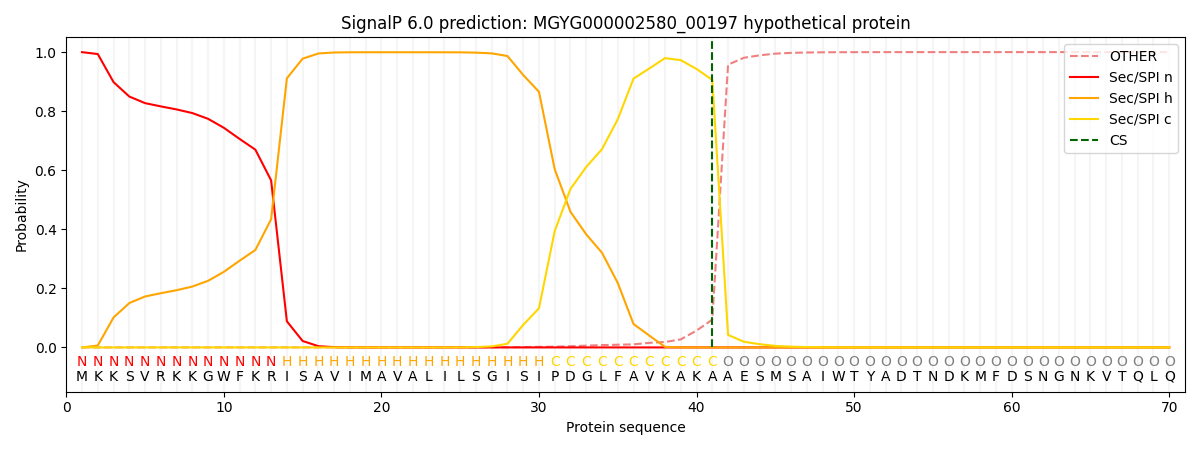
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000390 | 0.998611 | 0.000415 | 0.000223 | 0.000175 | 0.000144 |