You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002584_00169
You are here: Home > Sequence: MGYG000002584_00169
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Akkermansia sp900545155 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Verrucomicrobiales; Akkermansiaceae; Akkermansia; Akkermansia sp900545155 | |||||||||||
| CAZyme ID | MGYG000002584_00169 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Glycosyl hydrolase family 109 protein 1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 240298; End: 241758 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 71 | 478 | 7.2e-156 | 0.9924812030075187 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01408 | GFO_IDH_MocA | 5.64e-12 | 75 | 207 | 1 | 120 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| COG0673 | MviM | 6.52e-11 | 75 | 235 | 4 | 157 | Predicted dehydrogenase [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHV74239.1 | 1.24e-291 | 1 | 486 | 1 | 481 |
| QWP73299.1 | 1.24e-291 | 1 | 486 | 1 | 481 |
| QUY58196.1 | 1.24e-291 | 1 | 486 | 1 | 481 |
| QWP60889.1 | 1.24e-291 | 1 | 486 | 1 | 481 |
| QHV61872.1 | 1.24e-291 | 1 | 486 | 1 | 481 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6T2B_A | 2.85e-215 | 56 | 485 | 24 | 447 | Glycosidehydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila],6T2B_B Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila],6T2B_C Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila],6T2B_D Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila] |
| 2IXA_A | 7.51e-77 | 75 | 477 | 21 | 432 | A-zyme,N-acetylgalactosaminidase [Elizabethkingia meningoseptica],2IXB_A Crystal structure of N-ACETYLGALACTOSAMINIDASE in complex with GalNAC [Elizabethkingia meningoseptica] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B2UL75 | 4.56e-288 | 1 | 486 | 1 | 481 | Glycosyl hydrolase family 109 protein 1 OS=Akkermansia muciniphila (strain ATCC BAA-835 / DSM 22959 / JCM 33894 / BCRC 81048 / CCUG 64013 / CIP 107961 / Muc) OX=349741 GN=Amuc_0017 PE=3 SV=1 |
| B2UQL7 | 1.79e-222 | 1 | 485 | 1 | 472 | Glycosyl hydrolase family 109 protein 2 OS=Akkermansia muciniphila (strain ATCC BAA-835 / DSM 22959 / JCM 33894 / BCRC 81048 / CCUG 64013 / CIP 107961 / Muc) OX=349741 GN=Amuc_0920 PE=1 SV=1 |
| A3D6B7 | 2.04e-164 | 7 | 485 | 6 | 457 | Glycosyl hydrolase family 109 protein OS=Shewanella baltica (strain OS155 / ATCC BAA-1091) OX=325240 GN=Sbal_2793 PE=3 SV=1 |
| A6WQ58 | 2.89e-164 | 7 | 485 | 6 | 457 | Glycosyl hydrolase family 109 protein OS=Shewanella baltica (strain OS185) OX=402882 GN=Shew185_2813 PE=3 SV=1 |
| Q8ECL7 | 8.21e-164 | 7 | 485 | 6 | 457 | Alpha-N-acetylgalactosaminidase OS=Shewanella oneidensis (strain MR-1) OX=211586 GN=nagA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TAT
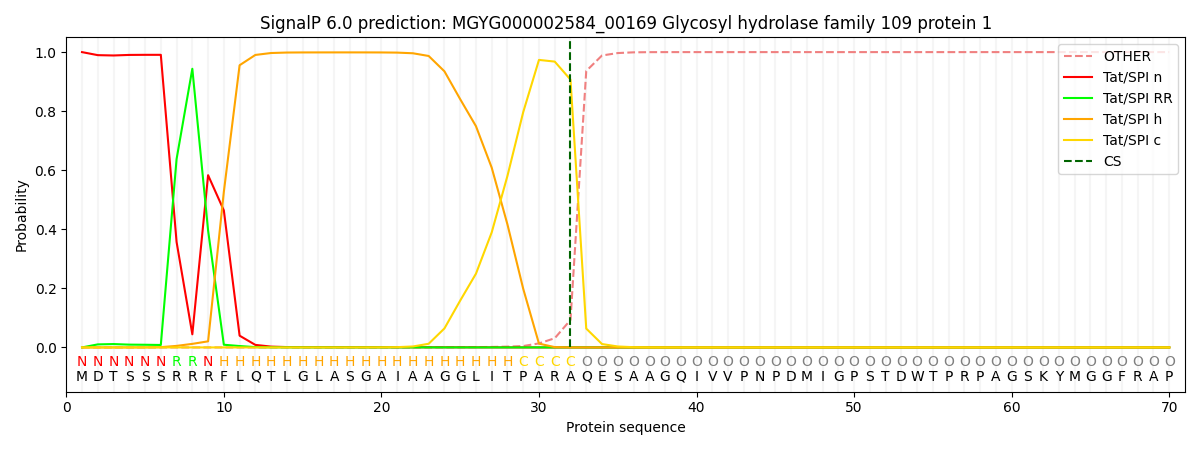
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 0.000000 | 0.999974 | 0.000005 | 0.000000 |
