You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002584_00415
You are here: Home > Sequence: MGYG000002584_00415
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Akkermansia sp900545155 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Verrucomicrobiales; Akkermansiaceae; Akkermansia; Akkermansia sp900545155 | |||||||||||
| CAZyme ID | MGYG000002584_00415 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3762; End: 6614 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 23 | 737 | 7.2e-69 | 0.726063829787234 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 1.79e-24 | 21 | 541 | 9 | 503 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 1.26e-20 | 27 | 346 | 15 | 296 | beta-D-glucuronidase; Provisional |
| pfam02837 | Glyco_hydro_2_N | 1.09e-08 | 27 | 210 | 4 | 159 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| PRK10340 | ebgA | 9.36e-07 | 291 | 454 | 281 | 455 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam02836 | Glyco_hydro_2_C | 3.96e-05 | 328 | 484 | 1 | 169 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT75482.1 | 0.0 | 27 | 941 | 26 | 931 |
| QRO25183.1 | 0.0 | 25 | 942 | 26 | 931 |
| BBE19496.1 | 0.0 | 25 | 941 | 27 | 941 |
| ATP55310.1 | 0.0 | 16 | 941 | 22 | 951 |
| AYB35766.1 | 0.0 | 23 | 938 | 48 | 964 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6SD0_A | 6.76e-17 | 25 | 451 | 40 | 443 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 6S6Z_A | 6.76e-17 | 25 | 451 | 39 | 442 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6U7I_A | 2.53e-15 | 116 | 466 | 62 | 426 | Faecalibacteriumprausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii],6U7I_B Faecalibacterium prausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii],6U7I_C Faecalibacterium prausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii],6U7I_D Faecalibacterium prausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii] |
| 5C70_A | 1.65e-13 | 25 | 478 | 21 | 453 | Thestructure of Aspergillus oryzae beta-glucuronidase [Aspergillus oryzae],5C70_B The structure of Aspergillus oryzae beta-glucuronidase [Aspergillus oryzae] |
| 5C71_A | 3.93e-13 | 25 | 478 | 46 | 478 | Thestructure of Aspergillus oryzae a-glucuronidase complexed with glycyrrhetinic acid monoglucuronide [Aspergillus oryzae],5C71_B The structure of Aspergillus oryzae a-glucuronidase complexed with glycyrrhetinic acid monoglucuronide [Aspergillus oryzae],5C71_C The structure of Aspergillus oryzae a-glucuronidase complexed with glycyrrhetinic acid monoglucuronide [Aspergillus oryzae],5C71_D The structure of Aspergillus oryzae a-glucuronidase complexed with glycyrrhetinic acid monoglucuronide [Aspergillus oryzae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 3.70e-16 | 25 | 451 | 40 | 443 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| Q04F24 | 2.02e-09 | 27 | 451 | 50 | 459 | Beta-galactosidase OS=Oenococcus oeni (strain ATCC BAA-331 / PSU-1) OX=203123 GN=lacZ PE=3 SV=1 |
| O33815 | 1.76e-08 | 295 | 456 | 273 | 445 | Beta-galactosidase OS=Staphylococcus xylosus OX=1288 GN=lacZ PE=3 SV=1 |
| Q0TKT1 | 6.92e-08 | 27 | 451 | 55 | 464 | Beta-galactosidase OS=Escherichia coli O6:K15:H31 (strain 536 / UPEC) OX=362663 GN=lacZ PE=3 SV=1 |
| Q8FKG6 | 6.92e-08 | 27 | 451 | 55 | 464 | Beta-galactosidase OS=Escherichia coli O6:H1 (strain CFT073 / ATCC 700928 / UPEC) OX=199310 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
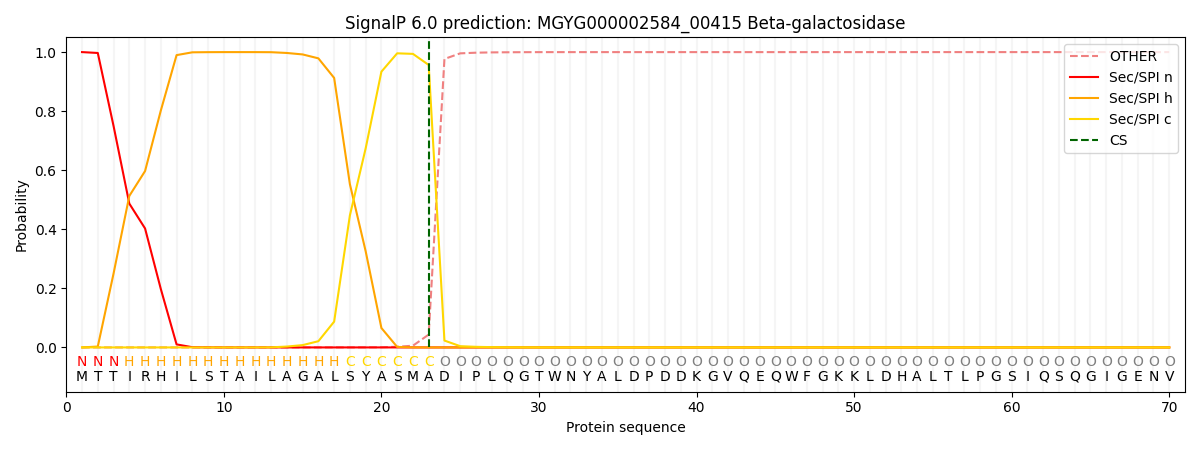
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000351 | 0.998836 | 0.000215 | 0.000212 | 0.000187 | 0.000161 |
