You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002586_01346
You are here: Home > Sequence: MGYG000002586_01346
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
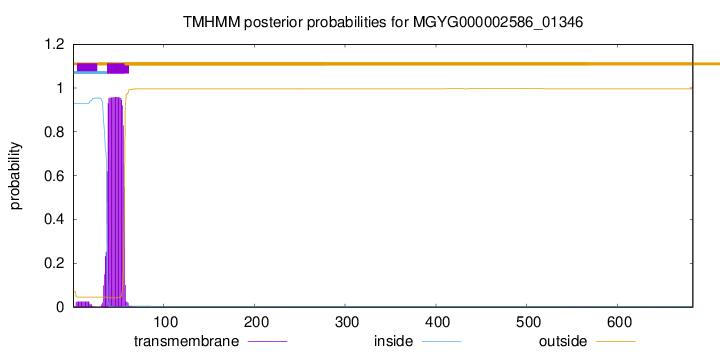
TMHMM annotations
Basic Information help
| Species | CAG-145 sp002320005 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Peptostreptococcales; Anaerovoracaceae; CAG-145; CAG-145 sp002320005 | |||||||||||
| CAZyme ID | MGYG000002586_01346 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3816; End: 5867 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 2.07e-24 | 218 | 438 | 46 | 245 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| cd14256 | Dockerin_I | 8.73e-17 | 620 | 676 | 1 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| pfam00404 | Dockerin_1 | 1.17e-13 | 621 | 675 | 1 | 55 | Dockerin type I repeat. The dockerin repeat is the binding partner of the cohesin domain pfam00963. The cohesin-dockerin interaction is the crucial interaction for complex formation in the cellulosome. The dockerin repeats, each bearing homology to the EF-hand calcium-binding loop bind calcium. |
| COG4991 | YraI | 8.17e-11 | 42 | 136 | 9 | 106 | Uncharacterized conserved protein YraI [Function unknown]. |
| pfam08239 | SH3_3 | 2.00e-10 | 72 | 122 | 1 | 53 | Bacterial SH3 domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBK77705.1 | 3.65e-129 | 71 | 657 | 47 | 847 |
| ACR71571.1 | 5.07e-129 | 74 | 656 | 53 | 664 |
| QRV21758.1 | 1.01e-120 | 70 | 647 | 44 | 749 |
| ADL03932.1 | 1.01e-120 | 70 | 647 | 44 | 749 |
| AEN96074.1 | 4.60e-116 | 57 | 657 | 117 | 735 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2KRS_A | 2.10e-08 | 67 | 124 | 4 | 61 | SolutionNMR structure of SH3 domain from CPF_0587 (fragment 415-479) from Clostridium perfringens. Northeast Structural Genomics Consortium (NESG) Target CpR74A. [Clostridium perfringens] |
| 4PI7_A | 2.18e-08 | 289 | 424 | 90 | 212 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI8_A | 1.30e-07 | 289 | 424 | 90 | 212 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 6FXO_A | 2.08e-07 | 327 | 438 | 143 | 244 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
| 2KQ8_A | 3.18e-07 | 70 | 122 | 8 | 60 | ChainA, Cell wall hydrolase [[Bacillus thuringiensis] serovar konkukian] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q931U5 | 4.70e-06 | 327 | 438 | 1147 | 1248 | Bifunctional autolysin OS=Staphylococcus aureus (strain Mu50 / ATCC 700699) OX=158878 GN=atl PE=1 SV=2 |
| Q99V41 | 4.70e-06 | 327 | 438 | 1147 | 1248 | Bifunctional autolysin OS=Staphylococcus aureus (strain N315) OX=158879 GN=atl PE=1 SV=1 |
| A7X0T9 | 4.71e-06 | 327 | 438 | 1154 | 1255 | Bifunctional autolysin OS=Staphylococcus aureus (strain Mu3 / ATCC 700698) OX=418127 GN=atl PE=3 SV=2 |
| P0C5Z8 | 4.71e-06 | 327 | 438 | 1154 | 1255 | Bifunctional autolysin OS=Staphylococcus aureus OX=1280 GN=atl PE=1 SV=1 |
| Q2FZK7 | 4.71e-06 | 327 | 438 | 1155 | 1256 | Bifunctional autolysin OS=Staphylococcus aureus (strain NCTC 8325 / PS 47) OX=93061 GN=atl PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
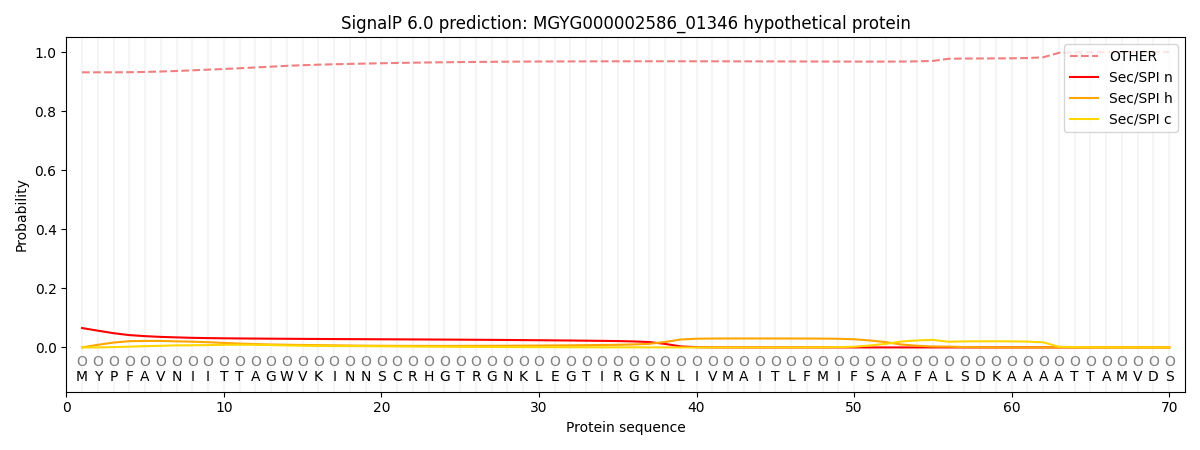
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.936830 | 0.058466 | 0.004393 | 0.000091 | 0.000045 | 0.000163 |