You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002592_00421
You are here: Home > Sequence: MGYG000002592_00421
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes sp902388705 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes sp902388705 | |||||||||||
| CAZyme ID | MGYG000002592_00421 | |||||||||||
| CAZy Family | GH147 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 141773; End: 143575 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH147 | 260 | 597 | 9.6e-43 | 0.3872549019607843 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3934 | COG3934 | 0.007 | 93 | 219 | 26 | 148 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL00993.1 | 0.0 | 1 | 599 | 1 | 599 |
| BBL08918.1 | 0.0 | 1 | 599 | 1 | 599 |
| BBL11710.1 | 0.0 | 1 | 599 | 1 | 599 |
| BAV09036.1 | 1.18e-45 | 2 | 596 | 4 | 531 |
| QXD15437.1 | 1.83e-44 | 46 | 534 | 28 | 475 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3PZ9_A | 6.56e-06 | 113 | 218 | 80 | 188 | Nativestructure of endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZG_A I222 crystal form of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZI_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with beta-D-glucose [Thermotoga petrophila RKU-1],3PZM_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules [Thermotoga petrophila RKU-1],3PZN_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with citrate and glycerol [Thermotoga petrophila RKU-1],3PZO_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with three maltose molecules [Thermotoga petrophila RKU-1],3PZQ_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with maltose and glycerol [Thermotoga petrophila RKU-1] |
| 6TN6_A | 9.09e-06 | 113 | 218 | 66 | 174 | X-raystructure of the endo-beta-1,4-mannanase from Thermotoga petrophila [Thermotoga petrophila RKU-1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q7Y223 | 1.49e-07 | 48 | 223 | 45 | 220 | Mannan endo-1,4-beta-mannosidase 2 OS=Arabidopsis thaliana OX=3702 GN=MAN2 PE=2 SV=1 |
| Q0DCM5 | 2.48e-06 | 116 | 242 | 114 | 241 | Mannan endo-1,4-beta-mannosidase 6 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN6 PE=2 SV=2 |
| Q9M0H6 | 5.63e-06 | 99 | 219 | 83 | 215 | Mannan endo-1,4-beta-mannosidase 5 OS=Arabidopsis thaliana OX=3702 GN=MAN5 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
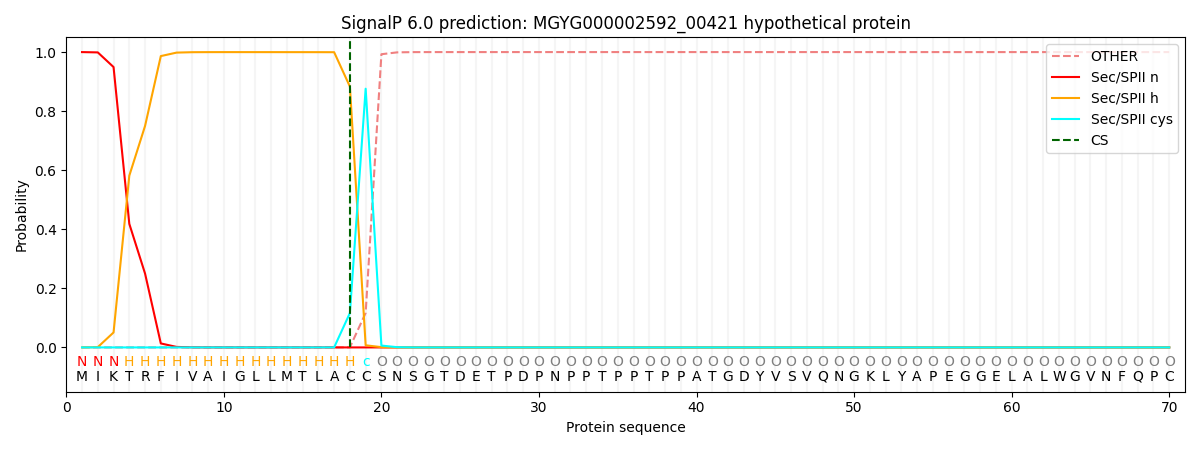
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000036 | 1.000018 | 0.000000 | 0.000000 | 0.000000 |
