You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002592_01141
You are here: Home > Sequence: MGYG000002592_01141
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes sp902388705 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes sp902388705 | |||||||||||
| CAZyme ID | MGYG000002592_01141 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 22207; End: 25050 Strand: - | |||||||||||
Full Sequence Download help
| MKLKFLLALL LAAFGTGLAH AGKLPVVKTP RPEKELYNVF KNPDERHRPY VRWWWNGSRV | 60 |
| DEREILRELD LMREAGIGGV EINTIQFPDQ TADTVGCAAL TWLSDEWIHM VNVAADGCRE | 120 |
| RGMTCDIIVG SGWPFGAEYL GPDEQVQMLF PVTIDVGEGK FTISRDEVLR MADARIANPR | 180 |
| SNPDKELMFI RLMPKNVEEF TPGVSYDGQA GNGQITVDVP QGEHVLYFFV KLSGYSRVIL | 240 |
| GAPGASGPVV NHLDGKAVGR YLDKFSDAMH FTRGKLKGEI RAAFCDSFEL EGDNWTPGMF | 300 |
| AEFERRMGYS LDPYLPYVFQ KTGAMGEPVR EAYGSNFSVK VTREVVERVR HDFWHVQMQL | 360 |
| FKENFIDVYN AWCHRNGLKS RIQAYGHQLH PIEASMYIDI PECESWIHDG IGRVMNPNEY | 420 |
| LSGRGYSMVN KFVSSGALLA GRPVVSCEEQ TNVGNIFQTT LEEVKVTGDR SNLSGVNHSV | 480 |
| LHGFNYSPRQ KDFFGWIQFG TYFNENNTWW PYVRSWTDYK ARVSALLQNS VYQADIAILP | 540 |
| PLEDLWSIHG MQRDPYPGVT HPAYANDLWE AVMQSGNGCD YVSENIIRQS SVGKGCIRFG | 600 |
| ARSYNTLLLM EVESLEPETA ERIASFVAAG GRVIAIGKVP CKGLGFKDAD AKDARVKAII | 660 |
| DRTVAAHPDR FIRVDAPAGD ILGWYRGLQA EYGLTPYAKI SAPDRFLSMN YYKSGGKDIY | 720 |
| FLVNSSIERT IQTRVEFPAA VAAKQAWLWN AETGERHMLG VQNGAIELCL TPAEAKFIVF | 780 |
| EKDCSGAMLP APAPRNANPV ALTGIWDVKA THHVDKSVKE FRLTDLVDLH SLPFPWLQSF | 840 |
| AGEIEYSRTV EIEDPAAFHT LDAGLTHNGL TELFVNGESA GVRWYGARTF DVGGMLRKGE | 900 |
| NRITIRVTTV LTNYAKARAA DTPTAARWEW AQRLNKELGL RGPVTIY | 947 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 36 | 946 | 2.2e-172 | 0.9951456310679612 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 1.98e-50 | 194 | 760 | 350 | 870 | alpha-L-rhamnosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL12421.1 | 0.0 | 1 | 947 | 1 | 947 |
| BBL09627.1 | 0.0 | 1 | 947 | 1 | 947 |
| BBL01754.1 | 0.0 | 1 | 947 | 1 | 947 |
| BCG53799.1 | 0.0 | 36 | 947 | 1 | 920 |
| QNL48206.1 | 3.18e-268 | 11 | 944 | 14 | 942 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q2F_A | 1.24e-31 | 194 | 913 | 409 | 1100 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
| 5MQM_A | 1.79e-15 | 215 | 946 | 386 | 1098 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 4.06e-15 | 215 | 946 | 386 | 1098 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNA8 | 2.46e-38 | 3 | 781 | 5 | 739 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
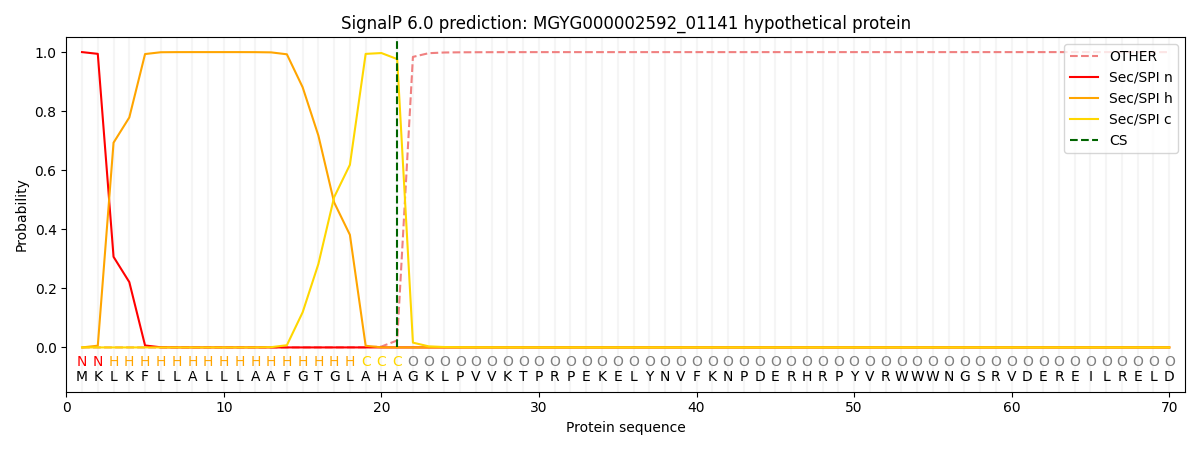
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000241 | 0.999138 | 0.000161 | 0.000158 | 0.000140 | 0.000130 |
