You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002595_01132
You are here: Home > Sequence: MGYG000002595_01132
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
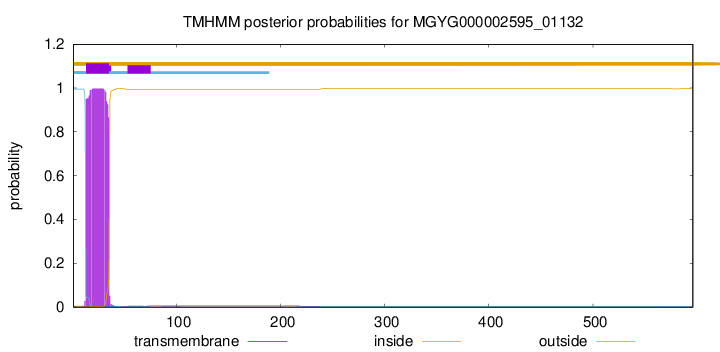
TMHMM annotations
Basic Information help
| Species | CAG-83 sp902388735 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; CAG-83; CAG-83 sp902388735 | |||||||||||
| CAZyme ID | MGYG000002595_01132 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1506; End: 3296 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 2.77e-32 | 192 | 394 | 38 | 231 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| cd14256 | Dockerin_I | 9.60e-11 | 533 | 588 | 1 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| smart00047 | LYZ2 | 3.37e-10 | 304 | 382 | 52 | 130 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| pfam00404 | Dockerin_1 | 1.69e-09 | 534 | 588 | 1 | 56 | Dockerin type I repeat. The dockerin repeat is the binding partner of the cohesin domain pfam00963. The cohesin-dockerin interaction is the crucial interaction for complex formation in the cellulosome. The dockerin repeats, each bearing homology to the EF-hand calcium-binding loop bind calcium. |
| smart00287 | SH3b | 3.93e-04 | 39 | 109 | 4 | 61 | Bacterial SH3 domain homologues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCK79746.1 | 1.09e-286 | 1 | 596 | 1 | 588 |
| QWT55084.1 | 4.05e-59 | 41 | 568 | 48 | 577 |
| QWT53775.1 | 3.95e-50 | 63 | 560 | 59 | 571 |
| AEN96074.1 | 4.08e-50 | 43 | 571 | 132 | 736 |
| ABX43817.1 | 2.24e-45 | 41 | 430 | 755 | 1189 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FXO_A | 2.87e-15 | 260 | 381 | 95 | 217 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
| 6FXP_A | 1.13e-14 | 258 | 381 | 102 | 219 | ChainA, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50],6FXP_B Chain B, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50] |
| 6U0O_B | 2.06e-14 | 258 | 381 | 132 | 249 | ChainB, LYZ2 domain-containing protein [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 4PI7_A | 4.14e-12 | 260 | 381 | 87 | 201 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI8_A | 2.52e-11 | 260 | 381 | 87 | 201 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P39848 | 6.01e-18 | 195 | 399 | 672 | 868 | Beta-N-acetylglucosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=lytD PE=1 SV=1 |
| Q6GAG0 | 2.03e-13 | 260 | 381 | 1101 | 1223 | Bifunctional autolysin OS=Staphylococcus aureus (strain MSSA476) OX=282459 GN=atl PE=3 SV=1 |
| Q8NX96 | 2.04e-13 | 260 | 381 | 1107 | 1229 | Bifunctional autolysin OS=Staphylococcus aureus (strain MW2) OX=196620 GN=atl PE=3 SV=1 |
| Q5HH31 | 2.68e-13 | 260 | 381 | 1107 | 1229 | Bifunctional autolysin OS=Staphylococcus aureus (strain COL) OX=93062 GN=atl PE=3 SV=1 |
| Q931U5 | 2.68e-13 | 260 | 381 | 1099 | 1221 | Bifunctional autolysin OS=Staphylococcus aureus (strain Mu50 / ATCC 700699) OX=158878 GN=atl PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002228 | 0.996538 | 0.000502 | 0.000260 | 0.000227 | 0.000223 |