You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002596_01591
You are here: Home > Sequence: MGYG000002596_01591
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Mediterraneibacter sp900555215 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Mediterraneibacter; Mediterraneibacter sp900555215 | |||||||||||
| CAZyme ID | MGYG000002596_01591 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6987; End: 10907 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH95 | 450 | 1101 | 4.9e-50 | 0.8379501385041551 |
| CBM32 | 318 | 445 | 5.7e-17 | 0.8629032258064516 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 1.24e-12 | 305 | 445 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 7.27e-09 | 135 | 267 | 1 | 120 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam07532 | Big_4 | 6.89e-08 | 1165 | 1219 | 5 | 57 | Bacterial Ig-like domain (group 4). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| cd00057 | FA58C | 3.38e-07 | 309 | 431 | 16 | 129 | Substituted updates: Jan 31, 2002 |
| smart00231 | FA58C | 4.64e-04 | 326 | 431 | 29 | 124 | Coagulation factor 5/8 C-terminal domain, discoidin domain. Cell surface-attached carbohydrate-binding domain, present in eukaryotes and assumed to have horizontally transferred to eubacterial genomes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHW32096.1 | 1.51e-220 | 39 | 1237 | 42 | 1181 |
| QXE33659.1 | 2.01e-174 | 42 | 1145 | 70 | 952 |
| AWS40658.1 | 1.60e-171 | 31 | 1145 | 30 | 926 |
| BCB76148.1 | 4.22e-151 | 45 | 1145 | 1 | 879 |
| QKW17790.1 | 8.85e-81 | 455 | 1119 | 135 | 754 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8L7W8 | 6.39e-09 | 654 | 1077 | 384 | 792 | Alpha-L-fucosidase 2 OS=Arabidopsis thaliana OX=3702 GN=FUC95A PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
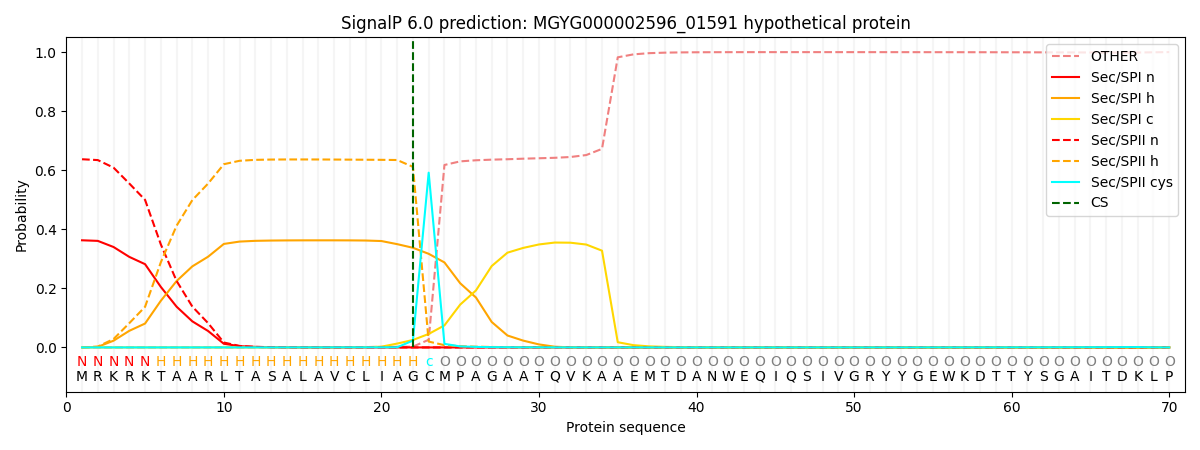
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000265 | 0.356380 | 0.642762 | 0.000245 | 0.000198 | 0.000138 |

