You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002596_02280
You are here: Home > Sequence: MGYG000002596_02280
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
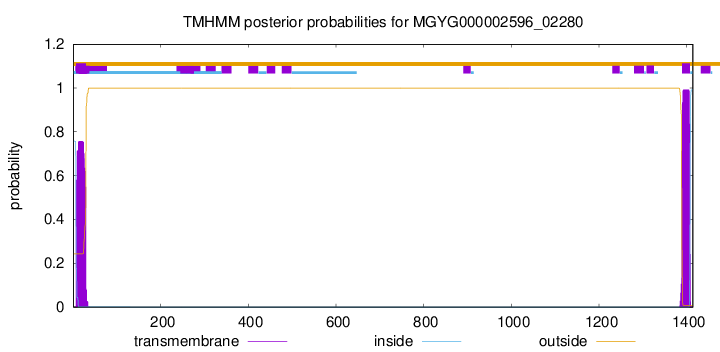
TMHMM annotations
Basic Information help
| Species | Mediterraneibacter sp900555215 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Mediterraneibacter; Mediterraneibacter sp900555215 | |||||||||||
| CAZyme ID | MGYG000002596_02280 | |||||||||||
| CAZy Family | CBM40 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3810; End: 8054 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 426 | 866 | 2.8e-91 | 0.935672514619883 |
| CBM40 | 228 | 399 | 9e-34 | 0.9217877094972067 |
| CBM40 | 49 | 202 | 2.9e-32 | 0.8770949720670391 |
| CBM32 | 1000 | 1130 | 1e-23 | 0.9435483870967742 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd15482 | Sialidase_non-viral | 2.58e-67 | 426 | 876 | 3 | 339 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| COG4409 | NanH | 7.58e-21 | 197 | 853 | 52 | 691 | Neuraminidase (sialidase) [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| pfam00754 | F5_F8_type_C | 8.48e-20 | 999 | 1130 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam02973 | Sialidase | 1.29e-13 | 38 | 201 | 7 | 179 | Sialidase, N-terminal domain. |
| pfam02973 | Sialidase | 6.69e-10 | 219 | 398 | 7 | 181 | Sialidase, N-terminal domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJA08933.1 | 4.98e-118 | 207 | 956 | 38 | 760 |
| QCQ04139.1 | 1.63e-116 | 209 | 969 | 158 | 928 |
| AIF44402.1 | 3.64e-115 | 214 | 954 | 31 | 757 |
| QIM63029.1 | 3.43e-114 | 245 | 884 | 56 | 694 |
| BAB40435.1 | 7.07e-114 | 225 | 1281 | 54 | 1138 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5TSP_A | 4.52e-101 | 416 | 874 | 3 | 448 | Crystalstructure of the catalytic domain of Clostridium perfringens neuraminidase (NanI) in complex with a CHES [Clostridium perfringens ATCC 13124],5TSP_B Crystal structure of the catalytic domain of Clostridium perfringens neuraminidase (NanI) in complex with a CHES [Clostridium perfringens ATCC 13124] |
| 2BF6_A | 7.67e-100 | 416 | 874 | 2 | 447 | AtomicResolution Structure of the bacterial sialidase NanI from Clostridium perfringens in complex with alpha-Sialic Acid (Neu5Ac). [Clostridium perfringens] |
| 2VK5_A | 8.41e-100 | 416 | 874 | 2 | 447 | TheStructure Of Clostridium Perfringens Nani Sialidase And Its Catalytic Intermediates [Clostridium perfringens],2VK6_A The Structure Of Clostridium Perfringens Nani Sialidase And Its Catalytic Intermediates [Clostridium perfringens],2VK7_A The Structure Of Clostridium Perfringens Nani Sialidase And Its Catalytic Intermediates [Clostridium perfringens],2VK7_B The Structure Of Clostridium Perfringens Nani Sialidase And Its Catalytic Intermediates [Clostridium perfringens] |
| 5KKY_A | 2.01e-84 | 405 | 862 | 12 | 472 | Structureof Streptococcus pneumonia NanA bound with inhibitor 9N3Neu5Ac2en [Streptococcus pneumoniae],5KKY_B Structure of Streptococcus pneumonia NanA bound with inhibitor 9N3Neu5Ac2en [Streptococcus pneumoniae] |
| 3H72_A | 2.19e-84 | 415 | 862 | 2 | 452 | Crystalstructure of Streptococcus pneumoniae D39 neuraminidase A precursor (NanA) in complex with NANA [Streptococcus pneumoniae R6],3H72_B Crystal structure of Streptococcus pneumoniae D39 neuraminidase A precursor (NanA) in complex with NANA [Streptococcus pneumoniae R6],3H73_A Crystal structure of Streptococcus pneumoniae D39 neuraminidase A precursor (NanA) in complex with DANA [Streptococcus pneumoniae R6],3H73_B Crystal structure of Streptococcus pneumoniae D39 neuraminidase A precursor (NanA) in complex with DANA [Streptococcus pneumoniae R6] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P29767 | 2.06e-105 | 175 | 984 | 165 | 935 | Sialidase OS=Clostridium septicum OX=1504 PE=3 SV=1 |
| P62576 | 1.42e-94 | 211 | 986 | 114 | 881 | Sialidase A OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=nanA PE=1 SV=1 |
| P62575 | 1.42e-94 | 211 | 986 | 114 | 881 | Sialidase A OS=Streptococcus pneumoniae OX=1313 GN=nanA PE=1 SV=1 |
| Q27701 | 4.78e-50 | 192 | 862 | 52 | 729 | Anhydrosialidase OS=Macrobdella decora OX=6405 PE=1 SV=1 |
| Q02834 | 4.06e-26 | 627 | 1132 | 177 | 643 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
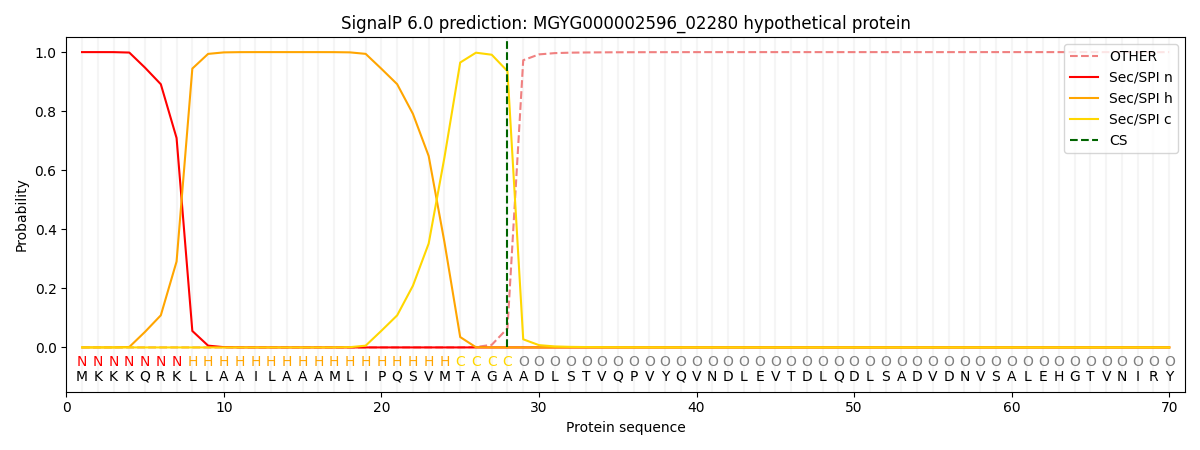
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000488 | 0.998476 | 0.000264 | 0.000287 | 0.000246 | 0.000219 |