You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002601_01550
You are here: Home > Sequence: MGYG000002601_01550
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-110 sp900549705 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; CAG-110; CAG-110 sp900549705 | |||||||||||
| CAZyme ID | MGYG000002601_01550 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-glucosidase BoGH3B | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 9771; End: 11657 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 135 | 351 | 1.5e-50 | 0.9722222222222222 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 2.04e-49 | 80 | 455 | 1 | 373 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| PRK15098 | PRK15098 | 9.43e-44 | 60 | 626 | 22 | 651 | beta-glucosidase BglX. |
| pfam00933 | Glyco_hydro_3 | 1.96e-32 | 81 | 382 | 1 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| pfam01915 | Glyco_hydro_3_C | 2.21e-22 | 421 | 621 | 2 | 214 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| PLN03080 | PLN03080 | 9.70e-12 | 198 | 627 | 155 | 628 | Probable beta-xylosidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QTE67436.1 | 2.48e-313 | 4 | 627 | 3 | 656 |
| QTE67433.1 | 1.98e-133 | 18 | 624 | 22 | 659 |
| AXH35175.1 | 5.45e-81 | 11 | 621 | 22 | 650 |
| ARJ06162.1 | 5.45e-81 | 11 | 628 | 22 | 654 |
| QXT42737.1 | 4.47e-75 | 32 | 621 | 34 | 676 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5M6G_A | 1.55e-57 | 62 | 621 | 40 | 612 | Crystalstructure Glucan 1,4-beta-glucosidase from Saccharopolyspora erythraea [Saccharopolyspora erythraea D] |
| 6JGL_A | 2.59e-51 | 62 | 625 | 9 | 605 | Crystalstructure of barley exohydrolaseI W434H mutant in complex with methyl 2-thio-beta-sophoroside [Hordeum vulgare subsp. vulgare],6JGN_A Crystal structure of barley exohydrolaseI W434H in complex with 4'-nitrophenyl thiolaminaribioside [Hordeum vulgare subsp. vulgare],6JGO_A Crystal structure of barley exohydrolaseI W434H mutant in complex with 4I,4III,4V-S-trithiocellohexaose [Hordeum vulgare subsp. vulgare],6JGP_A Crystal structure of barley exohydrolaseI W434H mutant in complex with methyl 6-thio-beta-gentiobioside. [Hordeum vulgare subsp. vulgare] |
| 6JGE_A | 3.56e-51 | 62 | 625 | 9 | 605 | Crystalstructure of barley exohydrolaseI W434A mutant in complex with methyl 2-thio-beta-sophoroside. [Hordeum vulgare subsp. vulgare],6K6V_A Crystal structure of barley exohydrolaseI W434A mutant in complex with methyl 6-thio-beta-gentiobioside [Hordeum vulgare subsp. vulgare],6KUF_A Crystal structure of barley exohydrolaseI W434A mutant in complex with glucose. [Hordeum vulgare subsp. vulgare],6L1J_A Crystal structure of barley exohydrolaseI W434A mutant in complex with 4'-nitrophenyl thiolaminaritrioside [Hordeum vulgare subsp. vulgare],6LBB_A Crystal structure of barley exohydrolaseI W434A mutant in complex with 4I,4III,4V-S-trithiocellohexaose [Hordeum vulgare subsp. vulgare] |
| 6JGQ_A | 4.89e-51 | 62 | 625 | 9 | 605 | Crystalstructure of barley exohydrolaseI W434Y mutant in complex with methyl 2-thio-beta-sophoroside. [Hordeum vulgare subsp. vulgare],6JGR_A Crystal structure of barley exohydrolaseI W434Y in complex with 4'-nitrophenyl thiolaminaribioside [Hordeum vulgare subsp. vulgare],6JGS_A Crystal structure of barley exohydrolaseI W434Y mutant in complex with 4I,4III,4V-S-trithiocellohexaose. [Hordeum vulgare subsp. vulgare],6JGT_A Crystal structure of barley exohydrolaseI W434Y mutant in complex with methyl 6-thio-beta-gentiobioside. [Hordeum vulgare subsp. vulgare] |
| 1LQ2_A | 6.04e-51 | 62 | 625 | 5 | 601 | Crystalstructure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with gluco-phenylimidazole [Hordeum vulgare],1X38_A crystal structure of barley beta-D-glucan glucohydrolase isoenzyme exo1 in complex with gluco-phenylimidazole [Hordeum vulgare],1X39_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme exo1 in complex with gluco-phenylimidazole [Hordeum vulgare] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q46684 | 8.16e-66 | 30 | 621 | 26 | 652 | Periplasmic beta-glucosidase/beta-xylosidase OS=Dickeya chrysanthemi OX=556 GN=bgxA PE=3 SV=1 |
| Q5BCC6 | 1.96e-60 | 71 | 624 | 47 | 616 | Beta-glucosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglC PE=1 SV=1 |
| Q2UFP8 | 1.96e-59 | 47 | 621 | 33 | 624 | Probable beta-glucosidase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=bglC PE=3 SV=2 |
| B8NGU6 | 6.41e-58 | 47 | 621 | 29 | 620 | Probable beta-glucosidase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bglC PE=3 SV=1 |
| A7LXU3 | 3.89e-47 | 77 | 593 | 43 | 617 | Beta-glucosidase BoGH3B OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02659 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
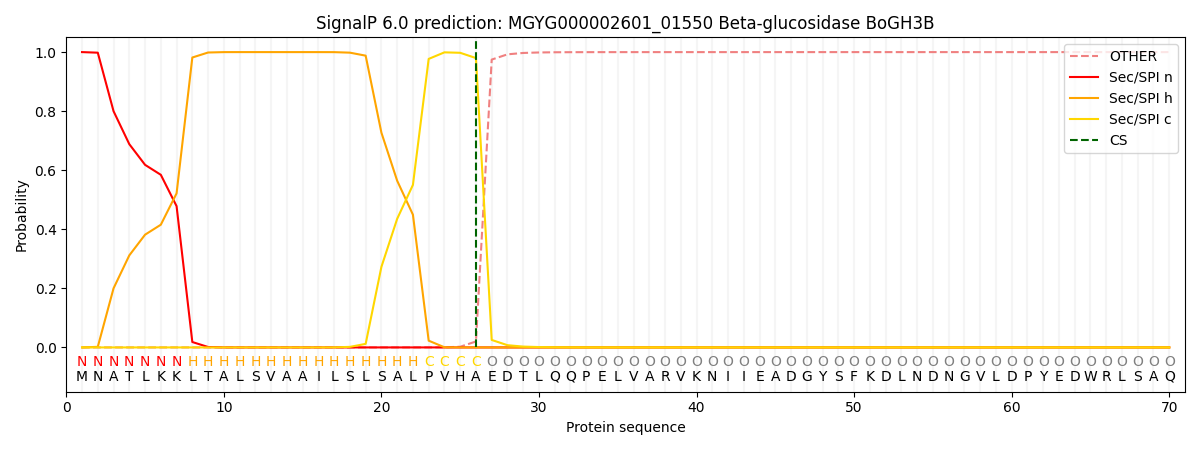
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000267 | 0.998975 | 0.000199 | 0.000196 | 0.000190 | 0.000163 |
